Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity
- PMID: 31668803
- PMCID: PMC6838684
- DOI: 10.1016/j.cell.2019.09.035
Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity
Abstract
Dendritic cells (DCs) play a critical role in orchestrating adaptive immune responses due to their unique ability to initiate T cell responses and direct their differentiation into effector lineages. Classical DCs have been divided into two subsets, cDC1 and cDC2, based on phenotypic markers and their distinct abilities to prime CD8 and CD4 T cells. While the transcriptional regulation of the cDC1 subset has been well characterized, cDC2 development and function remain poorly understood. By combining transcriptional and chromatin analyses with genetic reporter expression, we identified two principal cDC2 lineages defined by distinct developmental pathways and transcriptional regulators, including T-bet and RORγt, two key transcription factors known to define innate and adaptive lymphocyte subsets. These novel cDC2 lineages were characterized by distinct metabolic and functional programs. Extending our findings to humans revealed conserved DC heterogeneity and the presence of the newly defined cDC2 subsets in human cancer.
Keywords: ATAC-sequencing; T-bet; dendritic cells; myeloid cells; single-cell RNA-sequencing; transcriptional regulation.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures





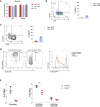
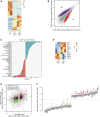



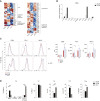
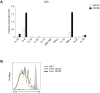


References
-
- Aliberti J., Schulz O., Pennington D.J., Tsujimura H., Reis e Sousa C., Ozato K., Sher A. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. - PubMed
-
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

