A composite immune signature parallels disease progression across T1D subjects
- PMID: 31671072
- PMCID: PMC6962023
- DOI: 10.1172/jci.insight.126917
A composite immune signature parallels disease progression across T1D subjects
Abstract
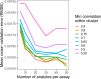
At diagnosis, most people with type 1 diabetes (T1D) produce measurable levels of endogenous insulin, but the rate at which insulin secretion declines is heterogeneous. To explain this heterogeneity, we sought to identify a composite signature predictive of insulin secretion, using a collaborative assay evaluation and analysis pipeline that incorporated multiple cellular and serum measures reflecting β cell health and immune system activity. The ability to predict decline in insulin secretion would be useful for patient stratification for clinical trial enrollment or therapeutic selection. Analytes from 12 qualified assays were measured in shared samples from subjects newly diagnosed with T1D. We developed a computational tool (DIFAcTO, Data Integration Flexible to Account for different Types of data and Outcomes) to identify a composite panel associated with decline in insulin secretion over 2 years following diagnosis. DIFAcTO uses multiple filtering steps to reduce data dimensionality, incorporates error estimation techniques including cross-validation and sensitivity analysis, and is flexible to assay type, clinical outcome, and disease setting. Using this novel analytical tool, we identified a panel of immune markers that, in combination, are highly associated with loss of insulin secretion. The methods used here represent a potentially novel process for identifying combined immune signatures that predict outcomes relevant for complex and heterogeneous diseases like T1D.
Keywords: Autoimmunity; Diabetes; Immunotherapy; Molecular pathology.
Conflict of interest statement
Figures




References
-
- Hao W, Gitelman S, DiMeglio LA, Boulware D, Greenbaum CJ, Type 1 Diabetes TrialNet Study Group Fall in C-peptide during first 4 years from diagnosis of type 1 diabetes: variable relation to age, HbA1c, and insulin dose. Diabetes Care. 2016;39(10):1664–1670. doi: 10.2337/dc16-0360. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

