Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape
- PMID: 31671153
- PMCID: PMC6957189
- DOI: 10.1371/journal.ppat.1008057
Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape
Abstract
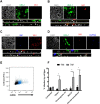
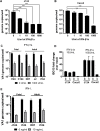
Human astroviruses (HAstV) are understudied positive-strand RNA viruses that cause gastroenteritis mostly in children and the elderly. Three clades of astroviruses, classic, MLB-type and VA-type have been reported in humans. One limitation towards a better understanding of these viruses has been the lack of a physiologically relevant cell culture model that supports growth of all clades of HAstV. Herein, we demonstrate infection of HAstV strains belonging to all three clades in epithelium-only human intestinal enteroids (HIE) isolated from biopsy-derived intestinal crypts. A detailed investigation of infection of VA1, a member of the non-canonical HAstV-VA/HMO clade, showed robust replication in HIE derived from different patients and from different intestinal regions independent of the cellular differentiation status. Flow cytometry and immunofluorescence analysis revealed that VA1 infects several cell types, including intestinal progenitor cells and mature enterocytes, in HIE cultures. RNA profiling of VA1-infected HIE uncovered that the host response to infection is dominated by interferon (IFN)-mediated innate immune responses. A comparison of the antiviral host response in non-transformed HIE and transformed human colon carcinoma Caco-2 cells highlighted significant differences between these cells, including an increased magnitude of the response in HIE. Additional studies confirmed the sensitivity of VA1 to exogenous IFNs, and indicated that the endogenous IFN response of HIE to curtail the growth of strains from all three clades. Genotypic variation in the permissiveness of different HIE lines to HAstV could be overcome by pharmacologic inhibition of JAK/STAT signaling. Collectively, our data identify HIE as a universal infection model for HAstV and an improved model of the intestinal epithelium to investigate enteric virus-host interactions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

