Post-Translational Deimination of Immunological and Metabolic Protein Markers in Plasma and Extracellular Vesicles of Naked Mole-Rat (Heterocephalus glaber)
- PMID: 31671738
- PMCID: PMC6862702
- DOI: 10.3390/ijms20215378
Post-Translational Deimination of Immunological and Metabolic Protein Markers in Plasma and Extracellular Vesicles of Naked Mole-Rat (Heterocephalus glaber)
Abstract
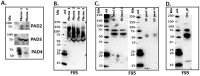
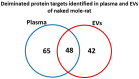
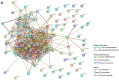
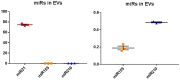
Naked mole-rats are long-lived animals that show unusual resistance to hypoxia, cancer and ageing. Protein deimination is an irreversible post-translational modification caused by the peptidylarginine deiminase (PAD) family of enzymes, which convert arginine into citrulline in target proteins. Protein deimination can cause structural and functional protein changes, facilitating protein moonlighting, but also leading to neo-epitope generation and effects on gene regulation. Furthermore, PADs have been found to regulate cellular release of extracellular vesicles (EVs), which are lipid-vesicles released from cells as part of cellular communication. EVs carry protein and genetic cargo and are indicative biomarkers that can be isolated from most body fluids. This study was aimed at profiling deiminated proteins in plasma and EVs of naked mole-rat. Key immune and metabolic proteins were identified to be post-translationally deiminated, with 65 proteins specific for plasma, while 42 proteins were identified to be deiminated in EVs only. Using protein-protein interaction network analysis, deiminated plasma proteins were found to belong to KEEG (Kyoto Encyclopedia of Genes and Genomes) pathways of immunity, infection, cholesterol and drug metabolism, while deiminated proteins in EVs were also linked to KEEG pathways of HIF-1 signalling and glycolysis. The mole-rat EV profiles showed a poly-dispersed population of 50-300 nm, similar to observations of human plasma. Furthermore, the EVs were assessed for three key microRNAs involved in cancer, inflammation and hypoxia. The identification of post-translational deimination of critical immunological and metabolic markers contributes to the current understanding of protein moonlighting functions, via post-translational changes, in the longevity and cancer resistance of naked mole-rats.
Keywords: extracellular vesicles (EVs); immunity; metabolism; miR155; miR210); microRNA (miR21; naked mole-rat (Heterocephalus glaber); peptidylarginine deiminases (PADs); protein deimination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Deimination Protein Profiles in Alligator mississippiensis Reveal Plasma and Extracellular Vesicle-Specific Signatures Relating to Immunity, Metabolic Function, and Gene Regulation.Front Immunol. 2020 Apr 28;11:651. doi: 10.3389/fimmu.2020.00651. eCollection 2020. Front Immunol. 2020. PMID: 32411128 Free PMC article.
-
Protein Deimination Signatures in Plasma and Plasma-EVs and Protein Deimination in the Brain Vasculature in a Rat Model of Pre-Motor Parkinson's Disease.Int J Mol Sci. 2020 Apr 15;21(8):2743. doi: 10.3390/ijms21082743. Int J Mol Sci. 2020. PMID: 32326590 Free PMC article.
-
Post-translational protein deimination signatures in sea lamprey (Petromyzon marinus) plasma and plasma-extracellular vesicles.Dev Comp Immunol. 2021 Dec;125:104225. doi: 10.1016/j.dci.2021.104225. Epub 2021 Aug 3. Dev Comp Immunol. 2021. PMID: 34358577
-
Peptidylarginine deiminases and deiminated proteins at the epidermal barrier.Exp Dermatol. 2018 Aug;27(8):852-858. doi: 10.1111/exd.13684. Epub 2018 Jun 29. Exp Dermatol. 2018. PMID: 29756256 Review.
-
Peptidylarginine deiminases and extracellular vesicles: prospective drug targets and biomarkers in central nervous system diseases and repair.Neural Regen Res. 2021 May;16(5):934-938. doi: 10.4103/1673-5374.297058. Neural Regen Res. 2021. PMID: 33229732 Free PMC article. Review.
Cited by
-
Deimination Protein Profiles in Alligator mississippiensis Reveal Plasma and Extracellular Vesicle-Specific Signatures Relating to Immunity, Metabolic Function, and Gene Regulation.Front Immunol. 2020 Apr 28;11:651. doi: 10.3389/fimmu.2020.00651. eCollection 2020. Front Immunol. 2020. PMID: 32411128 Free PMC article.
-
The Proteome and Citrullinome of Hippoglossus hippoglossus Extracellular Vesicles-Novel Insights into Roles of the Serum Secretome in Immune, Gene Regulatory and Metabolic Pathways.Int J Mol Sci. 2021 Jan 16;22(2):875. doi: 10.3390/ijms22020875. Int J Mol Sci. 2021. PMID: 33467210 Free PMC article.
-
Extracellular Vesicle Signatures and Post-Translational Protein Deimination in Purple Sea Urchin (Strongylocentrotus purpuratus) Coelomic Fluid-Novel Insights into Echinodermata Biology.Biology (Basel). 2021 Sep 3;10(9):866. doi: 10.3390/biology10090866. Biology (Basel). 2021. PMID: 34571743 Free PMC article.
-
Peptidylarginine Deiminase (PAD) and Post-Translational Protein Deimination-Novel Insights into Alveolata Metabolism, Epigenetic Regulation and Host-Pathogen Interactions.Biology (Basel). 2021 Feb 26;10(3):177. doi: 10.3390/biology10030177. Biology (Basel). 2021. PMID: 33653015 Free PMC article.
-
Low Cancer Incidence in Naked Mole-Rats May Be Related to Their Inability to Express the Warburg Effect.Front Physiol. 2022 May 4;13:859820. doi: 10.3389/fphys.2022.859820. eCollection 2022. Front Physiol. 2022. PMID: 35600297 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

