Autophagy in Cancer Cell Death
- PMID: 31671879
- PMCID: PMC6956186
- DOI: 10.3390/biology8040082
Autophagy in Cancer Cell Death
Abstract
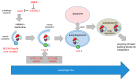
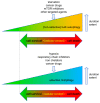
Autophagy has important functions in maintaining energy metabolism under conditions of starvation and to alleviate stress by removal of damaged and potentially harmful cellular components. Therefore, autophagy represents a pro-survival stress response in the majority of cases. However, the role of autophagy in cell survival and cell death decisions is highly dependent on its extent, duration, and on the respective cellular context. An alternative pro-death function of autophagy has been consistently observed in different settings, in particular, in developmental cell death of lower organisms and in drug-induced cancer cell death. This cell death is referred to as autophagic cell death (ACD) or autophagy-dependent cell death (ADCD), a type of cellular demise that may act as a backup cell death program in apoptosis-deficient tumors. This pro-death function of autophagy may be exerted either via non-selective bulk autophagy or excessive (lethal) removal of mitochondria via selective mitophagy, opening new avenues for the therapeutic exploitation of autophagy/mitophagy in cancer treatment.
Keywords: AT-101; autophagy; cancer; cell death; gossypol; mitochondria; mitophagy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

