Chronic Stress Induces Activity, Synaptic, and Transcriptional Remodeling of the Lateral Habenula Associated with Deficits in Motivated Behaviors
- PMID: 31672263
- PMCID: PMC6895430
- DOI: 10.1016/j.neuron.2019.09.005
Chronic Stress Induces Activity, Synaptic, and Transcriptional Remodeling of the Lateral Habenula Associated with Deficits in Motivated Behaviors
Abstract
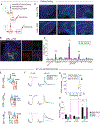
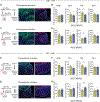
Chronic stress (CS) is a major risk factor for the development of depression. Here, we demonstrate that CS-induced hyperactivity in ventral tegmental area (VTA)-projecting lateral habenula (LHb) neurons is associated with increased passive coping (PC), but not anxiety or anhedonia. LHb→VTA neurons in mice with increased PC show increased burst and tonic firing as well as synaptic adaptations in excitatory inputs from the entopeduncular nucleus (EP). In vivo manipulations of EP→LHb or LHb→VTA neurons selectively alter PC and effort-related motivation. Conversely, dorsal raphe (DR)-projecting LHb neurons do not show CS-induced hyperactivity and are targeted indirectly by the EP. Using single-cell transcriptomics, we reveal a set of genes that can collectively serve as biomarkers to identify mice with increased PC and differentiate LHb→VTA from LHb→DR neurons. Together, we provide a set of biological markers at the level of genes, synapses, cells, and circuits that define a distinctive CS-induced behavioral phenotype.
Keywords: chronic stress; depression; dorsal raphe nucleus; entopeduncular nucleus; lateral habenula; ventral tegmental area.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- Agudelo LZ, Femenia T, Orhan F, Porsmyr-Palmertz M, Goiny M, Martinez-Redondo V, Correia JC, Izadi M, Bhat M, Schuppe-Koistinen I, et al. (2014). Skeletal muscle PGC-1α1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell 159, 33–45. - PubMed
-
- Bernard R, and Veh RW (2012). Individual neurons in the rat lateral habenular complex project mostly to the dopaminergic ventral tegmental area or to the serotonergic raphe nuclei. J. Comp. Neurol 520, 2545–2558. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

