Stochastic modeling of human papillomavirusearly promoter gene regulation
- PMID: 31672406
- PMCID: PMC6937396
- DOI: 10.1016/j.jtbi.2019.110057
Stochastic modeling of human papillomavirusearly promoter gene regulation
Abstract
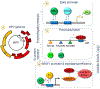
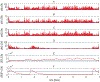
High risk forms of human papillomaviruses (HPVs) promote cancerous lesions and are implicated in almost all cervical cancer. Of particular relevance to cancer progression is regulation of the early promoter that controls gene expression in the initial phases of infection and can eventually lead to pre-cancer progression. Our goal was to develop a stochastic model to investigate the control mechanisms that regulate gene expression from the HPV early promoter. Our model integrates modules that account for transcriptional, post-transcriptional, translational and post-translational regulation of E1 and E2 early genes to form a functioning gene regulatory network. Each module consists of a set of biochemical steps whose stochastic evolution is governed by a chemical Master Equation and can be simulated using the Gillespie algorithm. To investigate the role of noise in gene expression, we compared our stochastic simulations with solutions to ordinary differential equations for the mean behavior of the system that are valid under the conditions of large molecular abundances and quasi-equilibrium for fast reactions. The model produced results consistent with known HPV biology. Our simulation results suggest that stochasticity plays a pivotal role in determining the dynamics of HPV gene expression. In particular, the combination of positive and negative feedback regulation generates stochastic bursts of gene expression. Analysis of the model reveals that regulation at the promoter affects burst amplitude and frequency, whereas splicing is more specialized to regulate burst frequency. Our results also suggest that splicing enhancers are a significant source of stochasticity in pre-mRNA abundance and that the number of viruses infecting the host cell represents a third important source of stochasticity in gene expression.
Keywords: Feedback regulation; RNA splicing; Stochastic bursting.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures










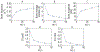
Similar articles
-
Stochasticity in transcriptional, splicing and translational regulations in time and frequency domains.Biosystems. 2022 Feb;212:104595. doi: 10.1016/j.biosystems.2021.104595. Epub 2022 Jan 10. Biosystems. 2022. PMID: 35026321
-
Stochastic modeling and numerical simulation of gene regulatory networks with protein bursting.J Theor Biol. 2017 May 21;421:51-70. doi: 10.1016/j.jtbi.2017.03.017. Epub 2017 Mar 21. J Theor Biol. 2017. PMID: 28341132
-
Periodic synchronization of isolated network elements facilitates simulating and inferring gene regulatory networks including stochastic molecular kinetics.BMC Bioinformatics. 2022 Jan 5;23(1):13. doi: 10.1186/s12859-021-04541-6. BMC Bioinformatics. 2022. PMID: 34986805 Free PMC article.
-
Regulatory elements in the viral genome.Virology. 2013 Oct;445(1-2):197-204. doi: 10.1016/j.virol.2013.04.035. Epub 2013 May 28. Virology. 2013. PMID: 23725692 Review.
-
Stochastic and delayed stochastic models of gene expression and regulation.Math Biosci. 2010 Jan;223(1):1-11. doi: 10.1016/j.mbs.2009.10.007. Epub 2009 Oct 31. Math Biosci. 2010. PMID: 19883665 Review.
References
-
- Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl.5):F55–70. - PubMed
-
- Bernard H-U. Gene expression of genital human papillomaviruses and considerations on potential antiviral approaches. Antivir Ther. 2002. December 1;7(4):219–37. Review. - PubMed
-
- Cobo F Human Papillomavirus Infections from the laboratory to clinical practice. 1st ed Woodhead Publishing; 2012.
-
- Johansson C, Schwartz S. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat Rev Microbiol. 2013. March 11;11(4):239–51. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

