Pseudouridinylation of mRNA coding sequences alters translation
- PMID: 31672910
- PMCID: PMC6859337
- DOI: 10.1073/pnas.1821754116
Pseudouridinylation of mRNA coding sequences alters translation
Abstract
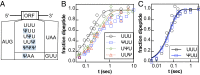
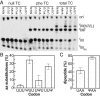
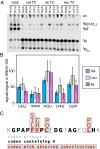
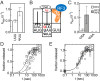
Chemical modifications of RNAs have long been established as key modulators of nonprotein-coding RNA structure and function in cells. There is a growing appreciation that messenger RNA (mRNA) sequences responsible for directing protein synthesis can also be posttranscriptionally modified. The enzymatic incorporation of mRNA modifications has many potential outcomes, including changing mRNA stability, protein recruitment, and translation. We tested how one of the most common modifications present in mRNA coding regions, pseudouridine (Ψ), impacts protein synthesis using a fully reconstituted bacterial translation system and human cells. Our work reveals that replacing a single uridine nucleotide with Ψ in an mRNA codon impedes amino acid addition and EF-Tu GTPase activation. A crystal structure of the Thermus thermophilus 70S ribosome with a tRNAPhe bound to a ΨUU codon in the A site supports these findings. We also find that the presence of Ψ can promote the low-level synthesis of multiple peptide products from a single mRNA sequence in the reconstituted translation system as well as human cells, and increases the rate of near-cognate Val-tRNAVal reacting on a ΨUU codon. The vast majority of Ψ moieties in mRNAs are found in coding regions, and our study suggests that one consequence of the ribosome encountering Ψ can be to modestly alter both translation speed and mRNA decoding.
Keywords: mRNA modification; pseudouridine; ribosome; translation.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
N1-Methylpseudouridine and pseudouridine modifications modulate mRNA decoding during translation.Nat Commun. 2024 Sep 16;15(1):8119. doi: 10.1038/s41467-024-51301-0. Nat Commun. 2024. PMID: 39284850 Free PMC article.
-
Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.Proc Natl Acad Sci U S A. 2020 Jul 14;117(28):16333-16338. doi: 10.1073/pnas.2004170117. Epub 2020 Jun 29. Proc Natl Acad Sci U S A. 2020. PMID: 32601241 Free PMC article.
-
N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.Nat Struct Mol Biol. 2016 Feb;23(2):110-5. doi: 10.1038/nsmb.3148. Epub 2016 Jan 11. Nat Struct Mol Biol. 2016. PMID: 26751643 Free PMC article.
-
Chemical modifications to mRNA nucleobases impact translation elongation and termination.Biophys Chem. 2022 Jun;285:106780. doi: 10.1016/j.bpc.2022.106780. Epub 2022 Feb 16. Biophys Chem. 2022. PMID: 35313212 Free PMC article. Review.
-
The ribosome in action: Tuning of translational efficiency and protein folding.Protein Sci. 2016 Aug;25(8):1390-406. doi: 10.1002/pro.2950. Epub 2016 Jun 8. Protein Sci. 2016. PMID: 27198711 Free PMC article. Review.
Cited by
-
From Antisense RNA to RNA Modification: Therapeutic Potential of RNA-Based Technologies.Biomedicines. 2021 May 14;9(5):550. doi: 10.3390/biomedicines9050550. Biomedicines. 2021. PMID: 34068948 Free PMC article. Review.
-
[Advances in mapping analysis of ribonucleic acid modifications through sequencing].Se Pu. 2024 Jul;42(7):632-645. doi: 10.3724/SP.J.1123.2023.12025. Se Pu. 2024. PMID: 38966972 Free PMC article. Review. Chinese.
-
Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.J Biol Chem. 2022 May;298(5):101869. doi: 10.1016/j.jbc.2022.101869. Epub 2022 Mar 26. J Biol Chem. 2022. PMID: 35346685 Free PMC article.
-
Extract2Chip-Bypassing Protein Purification in Drug Discovery Using Surface Plasmon Resonance.Biosensors (Basel). 2023 Oct 5;13(10):913. doi: 10.3390/bios13100913. Biosensors (Basel). 2023. PMID: 37887106 Free PMC article.
-
Transcriptome-wide mapping reveals a diverse dihydrouridine landscape including mRNA.PLoS Biol. 2022 May 24;20(5):e3001622. doi: 10.1371/journal.pbio.3001622. eCollection 2022 May. PLoS Biol. 2022. PMID: 35609439 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

