Clinically relevant molecular subtypes and genomic alteration-independent differentiation in gynecologic carcinosarcoma
- PMID: 31672974
- PMCID: PMC6823358
- DOI: 10.1038/s41467-019-12985-x
Clinically relevant molecular subtypes and genomic alteration-independent differentiation in gynecologic carcinosarcoma
Abstract
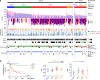
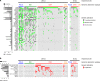
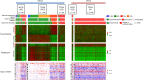
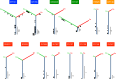
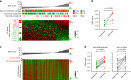
Carcinosarcoma (CS) of the uterus or ovary is a rare, aggressive and biphasic neoplasm composed of carcinoma and sarcoma elements. Previous genomic studies have identified the driver genes and genomic properties associated with CS. However, there is still no molecular subtyping scheme with clinical relevance for this disease. Here, we sequence 109 CS samples, focusing on 596 genes. We identify four molecular subtypes that resemble those observed in endometrial carcinoma: POLE-mutated, microsatellite instability, copy number high, and copy number low subtypes. These molecular subtypes are linked with DNA repair deficiencies, potential therapeutic strategies, and multiple clinicopathological features, including patient outcomes. Multi-regional comparative sequencing reveals genomic alteration-independent CS cell differentiation. Transcriptome and DNA methylome analyses confirm epithelial-mesenchymal transition as a mechanism of sarcoma differentiation. The current study thus provides therapeutic possibilities for CS as well as clues to understanding the molecular histogenic mechanism of its development.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

