UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts
- PMID: 31675358
- PMCID: PMC6853336
- DOI: 10.1371/journal.pgen.1008432
UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts
Abstract
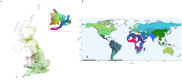
Human populations feature both discrete and continuous patterns of variation. Current analysis approaches struggle to jointly identify these patterns because of modelling assumptions, mathematical constraints, or numerical challenges. Here we apply uniform manifold approximation and projection (UMAP), a non-linear dimension reduction tool, to three well-studied genotype datasets and discover overlooked subpopulations within the American Hispanic population, fine-scale relationships between geography, genotypes, and phenotypes in the UK population, and cryptic structure in the Thousand Genomes Project data. This approach is well-suited to the influx of large and diverse data and opens new lines of inquiry in population-scale datasets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

