Target-Triggered Polymerization of Branched DNA Enables Enzyme-free and Fast Discrimination of Single-Base Changes
- PMID: 31675552
- PMCID: PMC6838547
- DOI: 10.1016/j.isci.2019.10.029
Target-Triggered Polymerization of Branched DNA Enables Enzyme-free and Fast Discrimination of Single-Base Changes
Abstract
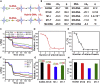
Single-base changes lead to important biological and biomedical implications; however, the discrimination of single-base changes from normal DNA always remains a grand challenge. Herein we developed a DNA recognition and amplification system based on artificial branched DNA, namely, target-triggered polymerization (TTP), to realize enzyme-free and fast discrimination of single-base changes. Branched DNA as monomers rapidly polymerized into DNA nanospheres only with the trigger of specific DNA. Our TTP system worked reliably over a wide range of conditions. Remarkably, our TTP system was capable of discriminating base-changing DNA from normal DNA, including distinguishing 1-4 nucleotide changes and positions of single base, which was attributed to the significant amplification of small differences in hybridization thermodynamics and kinetics. We further proposed a theoretical method for calculating the hybridization probability of nucleic acids, which performed highly consistent with experimental results.
Keywords: Nanostructure; Nanotechnology; Thermodynamics.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Abi A., Safavi A. Targeted detection of single-nucleotide variations: progress and promise. ACS Sens. 2019;4:792–807. - PubMed
-
- Ali M.M., Li F., Zhang Z., Zhang K., Kang D.K., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. - PubMed
-
- Bustamante C., Bryant Z., Smith S.B. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. - PubMed
-
- Chen Y., Xu J., Su J., Xiang Y., Yuan R., Chai Y. In situ hybridization chain reaction amplification for universal and highly sensitive electrochemiluminescent detection of DNA. Anal. Chem. 2012;84:7750–7755. - PubMed
LinkOut - more resources
Full Text Sources

