PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells
- PMID: 31676232
- PMCID: PMC7007862
- DOI: 10.1016/j.molcel.2019.10.008
PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells
Abstract
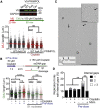
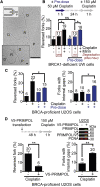
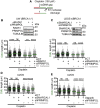
Acute treatment with replication-stalling chemotherapeutics causes reversal of replication forks. BRCA proteins protect reversed forks from nucleolytic degradation, and their loss leads to chemosensitivity. Here, we show that fork degradation is no longer detectable in BRCA1-deficient cancer cells exposed to multiple cisplatin doses, mimicking a clinical treatment regimen. This effect depends on increased expression and chromatin loading of PRIMPOL and is regulated by ATR activity. Electron microscopy and single-molecule DNA fiber analyses reveal that PRIMPOL rescues fork degradation by reinitiating DNA synthesis past DNA lesions. PRIMPOL repriming leads to accumulation of ssDNA gaps while suppressing fork reversal. We propose that cells adapt to repeated cisplatin doses by activating PRIMPOL repriming under conditions that would otherwise promote pathological reversed fork degradation. This effect is generalizable to other conditions of impaired fork reversal (e.g., SMARCAL1 loss or PARP inhibition) and suggests a new strategy to modulate cisplatin chemosensitivity by targeting the PRIMPOL pathway.
Keywords: ATR; BRCA; DNA damage; DNA replication; PRIMPOL; adaptive response; replication fork repriming; replication fork reversal; replication stress response; ssDNA gaps.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors decleare no competing interests.
Figures

References
-
- Antoniou A., Pharoah P.D., Narod S., Risch H.A., Eyfjord J.E., Hopper J.L., Loman N., Olsson H., Johannsson O., Borg A. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 2003;72:1117–1130. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

