Local Adaptation of Bacterial Symbionts within a Geographic Mosaic of Antibiotic Coevolution
- PMID: 31676475
- PMCID: PMC6881802
- DOI: 10.1128/AEM.01580-19
Local Adaptation of Bacterial Symbionts within a Geographic Mosaic of Antibiotic Coevolution
Abstract
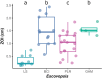
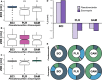
The geographic mosaic theory of coevolution (GMC) posits that coevolutionary dynamics go beyond local coevolution and are comprised of the following three components: geographic selection mosaics, coevolutionary hot spots, and trait remixing. It is unclear whether the GMC applies to bacteria, as horizontal gene transfer and cosmopolitan dispersal may violate theoretical assumptions. Here, we test key GMC predictions in an antibiotic-producing bacterial symbiont (genus Pseudonocardia) that protects the crops of neotropical fungus-farming ants (Apterostigma dentigerum) from a specialized pathogen (genus Escovopsis). We found that Pseudonocardia antibiotic inhibition of common Escovopsis pathogens was elevated in A. dentigerum colonies from Panama compared to those from Costa Rica. Furthermore, a Panama Canal Zone population of Pseudonocardia on Barro Colorado Island (BCI) was locally adapted, whereas two neighboring populations were not, consistent with a GMC-predicted selection mosaic and a hot spot of adaptation surrounded by areas of maladaptation. Maladaptation was shaped by incongruent Pseudonocardia-Escovopsis population genetic structure, whereas local adaptation was facilitated by geographic isolation on BCI after the flooding of the Panama Canal. Genomic assessments of antibiotic potential of 29 Pseudonocardia strains identified diverse and unique biosynthetic gene clusters in BCI strains despite low genetic diversity in the core genome. The strength of antibiotic inhibition was not correlated with the presence/absence of individual biosynthetic gene clusters or with parasite location. Rather, biosynthetic gene clusters have undergone selective sweeps, suggesting that the trait remixing dynamics conferring the long-term maintenance of antibiotic potency rely on evolutionary genetic changes within already-present biosynthetic gene clusters and not simply on the horizontal acquisition of novel genetic elements or pathways.IMPORTANCE Recently, coevolutionary theory in macroorganisms has been advanced by the geographic mosaic theory of coevolution (GMC), which considers how geography and local adaptation shape coevolutionary dynamics. Here, we test GMC in an ancient symbiosis in which the ant Apterostigma dentigerum cultivates fungi in an agricultural system analogous to human farming. The cultivars are parasitized by the fungus Escovopsis The ants maintain symbiotic actinobacteria with antibiotic properties that help combat Escovopsis infection. This antibiotic symbiosis has persisted for tens of millions of years, raising the question of how antibiotic potency is maintained over these time scales. Our study tests the GMC in a bacterial defensive symbiosis and in a multipartite symbiosis framework. Our results show that this multipartite symbiotic system conforms to the GMC and demonstrate that this theory is applicable in both microbes and indirect symbiont-symbiont interactions.
Keywords: coevolution; geographic mosaic theory of coevolution; secondary metabolism.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Margulis L. 1970. Origin of eukaryotic cells. Yale University Press, New Haven, CT.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

