Establishment of a novel human CIC-DUX4 sarcoma cell line, Kitra-SRS, with autocrine IGF-1R activation and metastatic potential to the lungs
- PMID: 31676869
- PMCID: PMC6825133
- DOI: 10.1038/s41598-019-52143-3
Establishment of a novel human CIC-DUX4 sarcoma cell line, Kitra-SRS, with autocrine IGF-1R activation and metastatic potential to the lungs
Erratum in
-
Author Correction: Establishment of a novel human CIC-DUX4 sarcoma cell line, Kitra-SRS, with autocrine IGF-1R activation and metastatic potential to the lungs.Sci Rep. 2020 Jan 15;10(1):684. doi: 10.1038/s41598-019-55752-0. Sci Rep. 2020. PMID: 31941891 Free PMC article.
Abstract
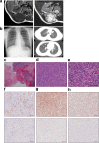
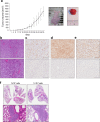
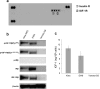
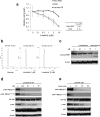
Approximately 60-70% of EWSR1-negative small blue round cell sarcomas harbour a rearrangement of CIC, most commonly CIC-DUX4. CIC-DUX4 sarcoma (CDS) is an aggressive and often fatal high-grade sarcoma appearing predominantly in children and young adults. Although cell lines and their xenograft models are essential tools for basic research and development of antitumour drugs, few cell lines currently exist for CDS. We successfully established a novel human CDS cell line designated Kitra-SRS and developed orthotopic tumour xenografts in nude mice. The CIC-DUX4 fusion gene in Kitra-SRS cells was generated by t(12;19) complex chromosomal rearrangements with an insertion of a chromosome segment including a DUX4 pseudogene component. Kitra-SRS xenografts were histologically similar to the original tumour and exhibited metastatic potential to the lungs. Kitra-SRS cells displayed autocrine activation of the insulin-like growth factor 1 (IGF-1)/IGF-1 receptor (IGF-1R) pathway. Accordingly, treatment with the IGF-1R inhibitor, linsitinib, attenuated Kitra-SRS cell growth and IGF-1-induced activation of IGF-1R/AKT signalling both in vitro and in vivo. Furthermore, upon screening 1134 FDA-approved drugs, the responses of Kitra-SRS cells to anticancer drugs appeared to reflect those of the primary tumour. Our model will be a useful modality for investigating the molecular pathology and therapy of CDS.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Yoshida A, et al. CIC-rearranged Sarcomas: A Study of 20 Cases and Comparisons With Ewing Sarcomas. Am. J. Surg. Pathol. 2016;40:313–323. - PubMed
-
- Sugita S, et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am. J. Surg. Pathol. 2014;38:1571–1576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

