Evasins: Tick Salivary Proteins that Inhibit Mammalian Chemokines
- PMID: 31679840
- PMCID: PMC7322545
- DOI: 10.1016/j.tibs.2019.10.003
Evasins: Tick Salivary Proteins that Inhibit Mammalian Chemokines
Abstract
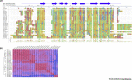
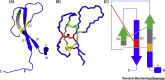
Ticks are hematophagous arachnids that parasitize mammals and other hosts, feeding on their blood. Ticks secrete numerous salivary factors that enhance host blood flow or suppress the host inflammatory response. The recruitment of leukocytes, a hallmark of inflammation, is regulated by chemokines, which activate chemokine receptors on the leukocytes. Ticks target this process by secreting glycoproteins called Evasins, which bind to chemokines and prevent leukocyte recruitment. This review describes the recent discovery of numerous Evasins produced by ticks, their classification into two structural and functional classes, and the efficacy of Evasins in animal models of inflammatory diseases. The review also proposes a standard nomenclature system for Evasins and discusses the potential of repurposing or engineering Evasins as therapeutic anti-inflammatory agents.
Keywords: Evasin; anti-inflammatory; binding protein; chemokine; protein family.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




References
-
- Murphy P.M. Chemokines and chemokine receptors. In: Rich R.R., editor. Clinical Immunology E-Book: Principles and Practice. 5th edn. Elsevier Health Sciences; 2018. pp. 157–170.
-
- Burns J.M. Comprehensive mapping of poxvirus vCCI chemokine-binding protein: expanded range of ligand interactions and unusual dissociation kinetics. J. Biol. Chem. 2002;277:2785–2789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

