Bacterial RNA Degradosomes: Molecular Machines under Tight Control
- PMID: 31679841
- PMCID: PMC6958999
- DOI: 10.1016/j.tibs.2019.10.002
Bacterial RNA Degradosomes: Molecular Machines under Tight Control
Abstract
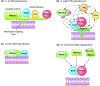
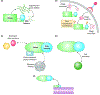
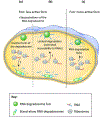
Bacterial RNA degradosomes are multienzyme molecular machines that act as hubs for post-transcriptional regulation of gene expression. The ribonuclease activities of these complexes require tight regulation, as they are usually essential for cell survival while potentially destructive. Recent studies have unveiled a wide variety of regulatory mechanisms including autoregulation, post-translational modifications, and protein compartmentalization. Recently, the subcellular organization of bacterial RNA degradosomes was found to present similarities with eukaryotic messenger ribonucleoprotein (mRNP) granules, membraneless compartments that are also involved in mRNA and protein storage and/or mRNA degradation. In this review, we present the current knowledge on the composition and targets of RNA degradosomes, the most recent developments regarding the regulation of these machineries, and their similarities with the eukaryotic mRNP granules.
Keywords: RNA degradation; RNA degradosome; RNA maturation; compartmentalization; mRNP granules; membraneless organelles; post-transcriptional regulation.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures
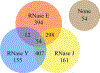
References
-
- Felden B, Paillard L (2017) When eukaryotes and prokaryotes look alike: the case of regulatory RNAs. FEMS Microbiol. Rev. 41, 624–639. - PubMed
-
- Aït-Bara S, Carpousis AJ (2015) RNA degradosomes in bacteria and chloroplasts: classification, distribution and evolution of RNase E homologs. Mol. Microbiol. 97, 1021–1135. - PubMed
-
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3’−5’ exoribonucleases. Cell 91, 457–66. - PubMed
-
- Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM (1994) Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76, 889–900. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

