Plant Reactome: a knowledgebase and resource for comparative pathway analysis
- PMID: 31680153
- PMCID: PMC7145600
- DOI: 10.1093/nar/gkz996
Plant Reactome: a knowledgebase and resource for comparative pathway analysis
Abstract
Plant Reactome (https://plantreactome.gramene.org) is an open-source, comparative plant pathway knowledgebase of the Gramene project. It uses Oryza sativa (rice) as a reference species for manual curation of pathways and extends pathway knowledge to another 82 plant species via gene-orthology projection using the Reactome data model and framework. It currently hosts 298 reference pathways, including metabolic and transport pathways, transcriptional networks, hormone signaling pathways, and plant developmental processes. In addition to browsing plant pathways, users can upload and analyze their omics data, such as the gene-expression data, and overlay curated or experimental gene-gene interaction data to extend pathway knowledge. The curation team actively engages researchers and students on gene and pathway curation by offering workshops and online tutorials. The Plant Reactome supports, implements and collaborates with the wider community to make data and tools related to genes, genomes, and pathways Findable, Accessible, Interoperable and Re-usable (FAIR).
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
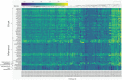
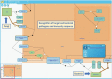

References
-
- Bolser D., Staines D.M., Pritchard E., Kersey P.. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016; 1374:115–140. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

