Identification and Analysis of the GASR Gene Family in Common Wheat (Triticum aestivum L.) and Characterization of TaGASR34, a Gene Associated With Seed Dormancy and Germination
- PMID: 31681420
- PMCID: PMC6813915
- DOI: 10.3389/fgene.2019.00980
Identification and Analysis of the GASR Gene Family in Common Wheat (Triticum aestivum L.) and Characterization of TaGASR34, a Gene Associated With Seed Dormancy and Germination
Abstract
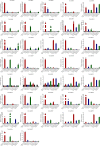
Seed dormancy and germination are important agronomic traits in wheat (Triticum aestivum L.) because they determine pre-harvest sprouting (PHS) resistance and thus affect grain production. These processes are regulated by Gibberellic Acid-Stimulated Regulator (GASR) genes. In this study, we identified 37 GASR genes in common wheat, which were designated TaGASR1-37. Moreover, we identified 40 pairs of paralogous genes, of which only one had a Ka/Ks value greater than 1, indicating that most TaGASR genes have undergone negative selection. Chromosomal location and duplication analysis revealed 25 pairs of segmentally duplicated genes and seven pairs of tandemly duplicated genes, suggesting that large-scale duplication events may have contributed to the expansion of TaGASR gene family. Microarray analysis of the expression of 18 TaGASR genes indicated that these genes play diverse roles in different biological processes. Using wheat varieties with contrasting seed dormancy phenotypes, we investigated the expression patterns of TaGASR genes and the corresponding seed germination index phenotypes in response to water imbibition, exogenous ABA and GA treatment, and low- and high-temperature treatment. Based on these data, we identified the TaGASR34 gene as potentially associated with seed dormancy and germination. Further, we used a SNP mutation of the TaGASR34 promoter (-16) to develop the CAPS marker GS34-7B, which was then used to validate the association of TaGASR34 with seed dormancy and germination by evaluating two natural populations across environments. Notably, the frequency of the high-dormancy GS34-7Bb allele was significantly lower than that of the low-dormancy GS34-7Ba allele, implying that the favorable GS34-7Bb allele has not previously been used in wheat breeding. These results provide valuable information for further functional analysis of TaGASR genes and present a useful gene and marker combination for future improvement of PHS resistance in wheat.
Keywords: ABA; GA; GASR; common wheat; seed dormancy.
Copyright © 2019 Cheng, Wang, Xu, Liu, Li, Xiao, Cao, Jiang, Min, Wang, Zhang, Chang, Lu and Ma.
Figures








References
-
- Alonso-Ramírez A., Rodríguez D., Reyes J., Jiménez J. A., Nicolás G., López-Climent M., et al. (2009). Evidence for a role of Gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 150, 1335–1344. 10.1104/pp.109.139352 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

