Exome Sequencing in BRCA1- and BRCA2-Negative Greek Families Identifies MDM1 and NBEAL1 as Candidate Risk Genes for Hereditary Breast Cancer
- PMID: 31681433
- PMCID: PMC6813924
- DOI: 10.3389/fgene.2019.01005
Exome Sequencing in BRCA1- and BRCA2-Negative Greek Families Identifies MDM1 and NBEAL1 as Candidate Risk Genes for Hereditary Breast Cancer
Abstract
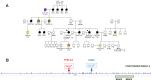
Approximately 10% of breast cancer (BC) cases are hereditary BC (HBC), with HBC most commonly encountered in the context of hereditary breast and ovarian cancer (HBOC) syndrome. Although thousands of loss-of-function (LoF) alleles in over 20 genes have been associated with HBC susceptibility, the genetic etiology of approximately 50% of cases remains unexplained, even when polygenic risk models are considered. We focused on one of the least-studied European populations and applied whole-exome sequencing (WES) to 52 individuals from 17 Greek HBOC families, in which at least one patient was negative for known HBC risk variants. Initial screening revealed pathogenic variants in known cancer genes, including BARD1:p.Trp91* detected in a cancer-free individual, and MEN1:p.Glu260Lys detected in a BC patient. Gene- and variant-based approaches were applied to exome data to identify candidate risk variants outside of known risk genes. Findings were verified in a collection of Canadian HBOC patients of European ancestry (FBRCAX), in an independent group of Canadian BC patients (CHUM-BC) and controls (CARTaGENE), as well as in individuals from The Cancer Genome Atlas (TCGA) and the UK Biobank (UKB). Rare LoF variants were uncovered in MDM1 and NBEAL1 in Greek and Canadian HBOC patients. We also report prioritized missense variants SETBP1:c.4129G > C and C7orf34:c.248C > T. These variants comprise promising candidates whose role in cancer pathogenicity needs to be explored further.
Keywords: Greek population; MDM1; NBEAL1; candidate risk variants; exome sequencing; hereditary breast cancer.
Copyright © 2019 Glentis, Dimopoulos, Rouskas, Ntritsos, Evangelou, Narod, Mes-Masson, Foulkes, Rivera, Tonin, Ragoussis and Dimas.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

