Embracing Ambiguity in the Taxonomic Classification of Microbiome Sequencing Data
- PMID: 31681437
- PMCID: PMC6811648
- DOI: 10.3389/fgene.2019.01022
Embracing Ambiguity in the Taxonomic Classification of Microbiome Sequencing Data
Abstract
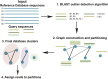
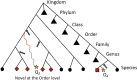
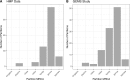
The advent of high throughput sequencing has enabled in-depth characterization of human and environmental microbiomes. Determining the taxonomic origin of microbial sequences is one of the first, and frequently only, analysis performed on microbiome samples. Substantial research has focused on the development of methods for taxonomic annotation, often making trade-offs in computational efficiency and classification accuracy. A side-effect of these efforts has been a reexamination of the bacterial taxonomy itself. Taxonomies developed prior to the genomic revolution captured complex relationships between organisms that went beyond uniform taxonomic levels such as species, genus, and family. Driven in part by the need to simplify computational workflows, the bacterial taxonomies used most commonly today have been regularized to fit within a standard seven taxonomic levels. Consequently, modern analyses of microbial communities are relatively coarse-grained. Few methods make classifications below the genus level, impacting our ability to capture biologically relevant signals. Here, we present ATLAS, a novel strategy for taxonomic annotation that uses significant outliers within database search results to group sequences in the database into partitions. These partitions capture the extent of taxonomic ambiguity within the classification of a sample. The ATLAS pipeline can be found on GitHub [https://github.com/shahnidhi/outlier_in_BLAST_hits]. We demonstrate that ATLAS provides similar annotations to phylogenetic placement methods, but with higher computational efficiency. When applied to human microbiome data, ATLAS is able to identify previously characterized taxonomic groupings, such as those in the class Clostridia and the genus Bacillus. Furthermore, the majority of partitions identified by ATLAS are at the subgenus level, replacing higher-level annotations with specific groups of species. These more precise partitions improve our detection power in determining differential abundance in microbiome association studies.
Keywords: 16S rRNA marker gene; classification; high-throughput sequencing; microbiome; taxonomy.
Copyright © 2019 Shah, Meisel and Pop.
Figures

References
-
- Bhandari V., Ahmod N. Z., Shah H. N., Gupta R. S. (2013). Molecular signatures for Bacillus species: demarcation of the Bacillus subtilis and Bacillus cereus clades in molecular terms and proposal to limit the placement of new species into the genus Bacillus. Int. J. Syst. Evol. Microbiol. 63, 2712–2726. 10.1099/ijs.0.048488-0 - DOI - PubMed
-
- Blondel V. D., Guillaume J.-L., Lambiotte R., Lefebvre E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008. 10.1088/1742-5468/2008/10/P10008 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

