Clinical, Cytogenetic, and Molecular Findings in Two Cases of Variant t(8;21) Acute Myeloid Leukemia (AML)
- PMID: 31681569
- PMCID: PMC6797852
- DOI: 10.3389/fonc.2019.01016
Clinical, Cytogenetic, and Molecular Findings in Two Cases of Variant t(8;21) Acute Myeloid Leukemia (AML)
Abstract
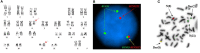
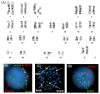
t(8;21)(q22;q22) is present in ~5-10% of patients with de novo acute myeloid leukemia (AML) and is associated with a better overall prognosis. Variants of the t(8;21) have been described in the literature, however, their clinical and prognostic significance has not been well-characterized. Molecular profiling of these cases has not previously been reported but may be useful in better defining the prognosis of this subset of patients. We present two cases of variant t(8;21) AML including clinical, cytogenetic, and molecular data.
Keywords: acute myeloid leukemia; core binding factor; cytogenetics; t(8;21); variant.
Copyright © 2019 Wilde, Cooper, Wang and Liu.
Figures
Similar articles
-
Comparison of cytogenetic and molecular genetic detection of t(8;21) and inv(16) in a prospective series of adults with de novo acute myeloid leukemia: a Cancer and Leukemia Group B Study.J Clin Oncol. 2001 May 1;19(9):2482-92. doi: 10.1200/JCO.2001.19.9.2482. J Clin Oncol. 2001. PMID: 11331327
-
Acute myeloid leukemia associated with variant t(8;21) detected by conventional cytogenetic and molecular studies: a report of four cases and review of the literature.Am J Clin Pathol. 2006 Feb;125(2):267-72. doi: 10.1309/8VJ4-V9PG-3TRJ-TLVH. Am J Clin Pathol. 2006. PMID: 16393685
-
A rare cytogenetic presentation of acute myeloid leukemia (AML-M2).Acta Med Iran. 2012;50(12):827-30. Acta Med Iran. 2012. PMID: 23456526 Review.
-
Acute Myeloid Leukemia With Recurrent Cytogenetic Abnormalities.Am J Clin Pathol. 2015 Jul;144(1):6-18. doi: 10.1309/AJCPI9C8UILYQTNS. Am J Clin Pathol. 2015. PMID: 26071458 Review.
-
Molecular and clinical advances in core binding factor primary acute myeloid leukemia: a paradigm for translational research in malignant hematology.Cancer Invest. 2000;18(8):768-80. doi: 10.3109/07357900009012209. Cancer Invest. 2000. PMID: 11107447 Review.
Cited by
-
Impact of additional genetic abnormalities at diagnosis of chronic myeloid leukemia for first-line imatinib-treated patients receiving proactive treatment intervention.Haematologica. 2023 Sep 1;108(9):2380-2395. doi: 10.3324/haematol.2022.282184. Haematologica. 2023. PMID: 36951160 Free PMC article.
-
Mitochondrial inhibitors: a new horizon in breast cancer therapy.Front Pharmacol. 2024 Jul 4;15:1421905. doi: 10.3389/fphar.2024.1421905. eCollection 2024. Front Pharmacol. 2024. PMID: 39027328 Free PMC article. Review.
-
Spatial-Temporal Genome Regulation in Stress-Response and Cell-Fate Change.Int J Mol Sci. 2023 Jan 31;24(3):2658. doi: 10.3390/ijms24032658. Int J Mol Sci. 2023. PMID: 36769000 Free PMC article. Review.
References
-
- Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. . Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. (2002) 100:4325–36. 10.1182/blood-2002-03-0772 - DOI - PubMed
-
- Prébet T, Boissel N, Reutenauer S, Thomas X, Delaunay J, Cahn J-Y, et al. . Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: a collaborative study of the french CBF-AML intergroup. J Clin Oncol. (2009) 27:4747–53. 10.1200/JCO.2008.21.0674 - DOI - PubMed
-
- Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. . The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. (1998) 92:2322–33. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

