Identification of a Five-Pseudogene Signature for Predicting Survival and Its ceRNA Network in Glioma
- PMID: 31681595
- PMCID: PMC6803554
- DOI: 10.3389/fonc.2019.01059
Identification of a Five-Pseudogene Signature for Predicting Survival and Its ceRNA Network in Glioma
Abstract
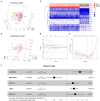
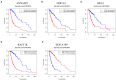
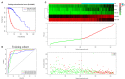
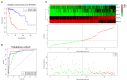
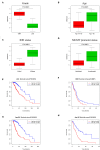
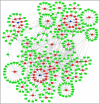
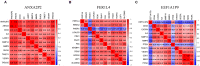
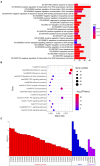
Background: Glioma is the most common primary brain tumor with a dismal prognosis. It is urgent to develop novel molecular biomarkers and conform to individualized schemes. Methods: Differentially expressed pseudogenes between low grade glioma (LGG) and glioblastoma multiforme (GBM) were identified in the training cohort. Least absolute shrinkage and selection operator (LASSO) regression and multivariate Cox proportional hazards regression analyses were used to select pseudogenes associated with prognosis of glioma. A risk signature was constructed based on the selected pseudogenes for predicting the survival of glioma patients. A pseudogene-miRNA-mRNA regulatory network was established and visualized using Cytoscape 3.5.1. Gene Oncology (GO) and signaling pathway analyses were performed on the targeted genes to investigate functional roles of the risk signature. Results: Five pseudogenes (ANXA2P2, EEF1A1P9, FER1L4, HILS1, and RAET1K) correlating with glioma survival were selected and used to establish a risk signature. Time-dependent receiver operating characteristic (ROC) curves revealed that the risk signature could accurately predict the 1, 3, and 5-year survival of glioma patients. GO and signaling pathway analyses showed that the risk signature was involved in regulation of proliferation, migration, angiogenesis, and apoptosis in glioma. Conclusions: In this study, a risk signature with five pseudogenes was constructed and shown to accurately predict 1-, 3-, and 5-year survival for glioma patient. The risk signature may serve as a potential target against glioma.
Keywords: ceRNA; glioma; nomogram; pseudogene; risk signature.
Copyright © 2019 Wang, Liu, Guan, Xiao, Zhao and Zhuang.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

