Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems
- PMID: 31681738
- PMCID: PMC6797904
- DOI: 10.3389/fbioe.2019.00248
Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems
Abstract
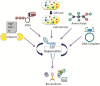
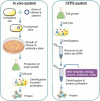
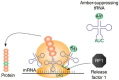
Cell-free protein synthesis (CFPS) system is a simple, rapid, and sensitive tool that is devoid of membrane-bound barriers, yet contains all the mandatory substrates, biomolecules, and machineries required for the synthesis of the desired proteins. It has the potential to overcome loopholes in the current in vivo production systems and is a promising tool in both basic and applied scientific research. It facilitates a simplified organization of desired experiments with a variety of reaction conditions, making CFPS a powerful tool in biological research. It has been used for the expansion of genetic code, assembly of viruses, and in metabolic engineering for production of toxic and complex proteins. Subsequently, CFPS systems have emerged as potent technology for high-throughput production of membrane proteins, enzymes, and therapeutics. The present review highlights the recent advances and uses of CFPS systems in biomedical, therapeutic, and biotechnological applications. Additionally, we highlight possible solutions to the potential biosafety issues that may be encountered while using CFPS technology.
Keywords: CFPS; biocontainment; high-throughput proteins; synthetic biology; therapeutics; virus-like particles.
Copyright © 2019 Khambhati, Bhattacharjee, Gohil, Braddick, Kulkarni and Singh.
Figures


Similar articles
-
Cell-Free Synthetic Biology: Engineering Beyond the Cell.Cold Spring Harb Perspect Biol. 2016 Dec 1;8(12):a023853. doi: 10.1101/cshperspect.a023853. Cold Spring Harb Perspect Biol. 2016. PMID: 27742731 Free PMC article. Review.
-
[Progress of cell-free protein synthesis system and its applications in pharmaceutical engineering - A review].Wei Sheng Wu Xue Bao. 2016 Mar 4;56(3):530-42. Wei Sheng Wu Xue Bao. 2016. PMID: 27382794 Review. Chinese.
-
Activation of Energy Metabolism through Growth Media Reformulation Enables a 24-Hour Workflow for Cell-Free Expression.ACS Synth Biol. 2020 Oct 16;9(10):2765-2774. doi: 10.1021/acssynbio.0c00283. Epub 2020 Sep 14. ACS Synth Biol. 2020. PMID: 32835484
-
Establishing a High-Yielding Cell-Free Protein Synthesis Platform Derived from Vibrio natriegens.ACS Synth Biol. 2018 Sep 21;7(9):2245-2255. doi: 10.1021/acssynbio.8b00252. Epub 2018 Sep 6. ACS Synth Biol. 2018. PMID: 30107122
-
[Cell-free protein synthetic system: progress and applications in biopharmaceutical engineering].Sheng Wu Gong Cheng Xue Bao. 2014 Oct;30(10):1491-503. Sheng Wu Gong Cheng Xue Bao. 2014. PMID: 25726574 Chinese.
Cited by
-
Influence of the mRNA initial region on protein production: a case study using recombinant detoxified pneumolysin as a model.Front Bioeng Biotechnol. 2024 Jan 8;11:1304965. doi: 10.3389/fbioe.2023.1304965. eCollection 2023. Front Bioeng Biotechnol. 2024. PMID: 38260740 Free PMC article.
-
Vaccine production and supply need a paradigm change.Can J Chem Eng. 2022 Aug;100(8):1670-1675. doi: 10.1002/cjce.24399. Epub 2022 May 4. Can J Chem Eng. 2022. PMID: 35572455 Free PMC article.
-
Promoting the production of challenging proteins via induced expression in CHO cells and modified cell-free lysates harboring T7 RNA polymerase and mutant eIF2α.Synth Syst Biotechnol. 2024 Mar 27;9(3):416-424. doi: 10.1016/j.synbio.2024.03.011. eCollection 2024 Sep. Synth Syst Biotechnol. 2024. PMID: 38601208 Free PMC article.
-
Solid-Phase Cell-Free Protein Synthesis and Its Applications in Biotechnology.Adv Biochem Eng Biotechnol. 2023;185:21-46. doi: 10.1007/10_2023_226. Adv Biochem Eng Biotechnol. 2023. PMID: 37306703
-
Recombinant Expression in Pichia pastoris System of Three Potent Kv1.3 Channel Blockers: Vm24, Anuroctoxin, and Ts6.J Fungi (Basel). 2022 Nov 17;8(11):1215. doi: 10.3390/jof8111215. J Fungi (Basel). 2022. PMID: 36422036 Free PMC article.
References
-
- Anand N. N., Mandal S., MacKenzie C. R., Sadowska J., Sigurskjold B., Young N. M., et al. . (1991). Bacterial expression and secretion of various single-chain Fv genes encoding proteins specific for a Salmonella serotype B O-antigen. J. Biol. Chem. 266, 21874–21879. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

