Intronic RNA: Ad'junk' mediator of post-transcriptional gene regulation
- PMID: 31682938
- PMCID: PMC7171924
- DOI: 10.1016/j.bbagrm.2019.194439
Intronic RNA: Ad'junk' mediator of post-transcriptional gene regulation
Abstract
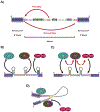
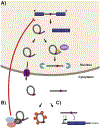
RNA splicing, the process through which intervening segments of noncoding RNA (introns) are excised from pre-mRNAs to allow for the formation of a mature mRNA product, has long been appreciated for its capacity to add complexity to eukaryotic proteomes. However, evidence suggests that the utility of this process extends beyond protein output and provides cells with a dynamic tool for gene regulation. In this review, we aim to highlight the role that intronic RNA plays in mediating specific splicing outcomes in pre-mRNA processing, as well as explore an emerging class of stable intronic sequences that have been observed to act in gene expression control. Building from underlying flexibility in both sequence and structure, intronic RNA provides mechanisms for post-transcriptional gene regulation that are amenable to the tissue and condition specific needs of eukaryotic cells. This article is part of a Special Issue entitled: RNA structure and splicing regulation edited by Francisco Baralle, Ravindra Singh and Stefan Stamm.
Copyright © 2019. Published by Elsevier B.V.
Figures
References
-
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. (2014) Nature 509: 575–581. doi:10.1038/nature13302. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

