Optimization of a Microplate Assay for Generating Listeria Monocytogenes, E. Coli O157:H7, and Salmonella Biofilms and Enzymatic Recovery for Enumeration
- PMID: 31684098
- PMCID: PMC6915590
- DOI: 10.3390/foods8110541
Optimization of a Microplate Assay for Generating Listeria Monocytogenes, E. Coli O157:H7, and Salmonella Biofilms and Enzymatic Recovery for Enumeration
Abstract
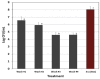
Biofilms enable the persistence of pathogens in food processing environments. Sanitizing agents are needed that are effective against pathogens entrapped in biofilms that are more difficult to inactivate than planktonic cells that are displaced and found on equipment surfaces. We examined conditions to develop, analyze, and enumerate the enhanced biofilms of three different foodborne pathogens assisted by fluorescence adherence assay and enzymatic detachment. We compared three different isomeric forms of fluorescent substrates that are readily taken up by bacterial cells based on carboxy-fluorescein diacetate (5-CFDA, 5,6-CFDA, 5,6-CFDA, SE). Biofilm-forming strains of Escherichia coli O157:H7 F4546 and Salmonella Montevideo FSIS 051 were identified using a microplate fluorescence assay defined previously for L. monocytogenes. Adherence levels were determined by differences in relative fluorescence units (RFU) as well as recovered bacterial cells. Multiple hydrolytic enzymes were examined for each representative pathogen for the most suitable enzyme for detachment and enumeration to confirm adherence data obtained by fluorescence assay. Cultures were grown overnight in microplates, incubated, washed and replenished with fresh sterile growth medium; this cycle was repeated for seven consecutive days to enrich for robust biofilms. Treatments were performed in triplicate and compared by one-way analysis of variance (ANOVA) to determine significant differences (p < 0.05).
Keywords: E. coli O157:H7; Listeria monocytogenes; Salmonella; biofilm; carboxyfluorescein diacetate; enzymes; microplate assay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Zhao X., Zhao F., Wang J., Zhong N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017;7:36670–36683. doi: 10.1039/C7RA02497E. - DOI
-
- Achinas S.C.N., Euverink G.J.W. A brief recap of microbial adhesion and biofilms. Appl. Sci. 2019;9:2801. doi: 10.3390/app9142801. - DOI
-
- Hinsa S.M., Espinosa-Urgel M., Ramos J.L., O’Toole G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

