Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy
- PMID: 31684890
- PMCID: PMC6829812
- DOI: 10.1186/s12879-019-4565-3
Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy
Abstract
Background: Endemic presence of Klebsiella pneumoniae resistant to carbapenem in Italy has been due principally to the clonal expansion of CC258 isolates; however, recent studies suggest an ongoing epidemiological change in this geographical area.
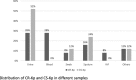
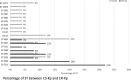
Methods: 50 K. pneumoniae strains, 25 carbapenem-resistant (CR-Kp) and 25 susceptible (CS-Kp), collected from march 2014 to march 2016 at the Laboratory of Bacteriology of the Paolo Giaccone Polyclinic University hospital of Palermo, Italy, were characterized for antibiotic susceptibility and fully sequenced by next generation sequencing (NGS) for the in silico analysis of resistome, virulome, multi-locus sequence typing (MLST) and core single nucleotide polymorphism (SNP) genotypes RESULTS: MLST in silico analysis of CR-Kp showed that 52% of isolates belonged to CC258, followed by ST395 (12%), ST307 (12%), ST392 (8%), ST348 (8%), ST405 (4%) and ST101 (4%). In the CS-Kp group, the most represented isolate was ST405 (20%), followed by ST392 and ST15 (12%), ST395, ST307 and ST1727 (8%). The in silico β-lactamase analysis of the CR-Kp group showed that the most detected gene was blaSHV (100%), followed by blaTEM (92%), blaKPC (88%), blaOXA (88%) and blaCTX-M (32%). The virulome analysis detected mrk operon in all studied isolates, and wzi-2 was found in three CR-Kp isolates (12%). Furthermore, the distribution of virulence genes encoding for the yersiniabactin system, its receptor fyuA and the aerobactin system did not show significant distribution differences between CR-Kp and CS-Kp, whereas the Klebsiella ferrous iron uptake system (kfuA, kfuB and kfuC genes), the two-component system kvgAS and the microcin E495 were significantly (p < 0.05) prevalent in the CS-Kp group compared to the CR-Kp group. Core SNP genotyping, correlating with the MLST data, allowed greater strain tracking and discrimination than MLST analysis.
Conclusions: Our data support the idea that an epidemiological change is ongoing in the Palermo area (Sicily, Italy). In addition, our analysis revealed the co-existence of antibiotic resistance and virulence factors in CR-Kp isolates; this characteristic should be considered for future genomic surveillance studies.
Keywords: Carbapenem-resistant Klebsiella pneumoniae; Virulence factors.
Conflict of interest statement
The author declare that they have no competing interests.
Figures
References
-
- World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 27 February 2017; 1–7.
-
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013; http://www.cdc.gov/drugresistance/threat-report-2013
-
- Department of Health and Department for Environment Food & Rural Affairs. UK Five Year Antimicrobial Resistance Strategy 2013 to 2018. 10 September 2013. https://www.gov.uk/government/publications/uk-5-year-antimicrobial-resis....
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;7(112):E3574–E3581. doi: 10.1073/pnas.1501049112. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical