Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration
- PMID: 31685987
- PMCID: PMC6940196
- DOI: 10.1038/s41556-019-0402-6
Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration
Abstract
Following severe or chronic liver injury, adult ductal cells (cholangiocytes) contribute to regeneration by restoring both hepatocytes and cholangiocytes. We recently showed that ductal cells clonally expand as self-renewing liver organoids that retain their differentiation capacity into both hepatocytes and ductal cells. However, the molecular mechanisms by which adult ductal-committed cells acquire cellular plasticity, initiate organoids and regenerate the damaged tissue remain largely unknown. Here, we describe that ductal cells undergo a transient, genome-wide, remodelling of their transcriptome and epigenome during organoid initiation and in vivo following tissue damage. TET1-mediated hydroxymethylation licences differentiated ductal cells to initiate organoids and activate the regenerative programme through the transcriptional regulation of stem-cell genes and regenerative pathways including the YAP-Hippo signalling. Our results argue in favour of the remodelling of genomic methylome/hydroxymethylome landscapes as a general mechanism by which differentiated cells exit a committed state in response to tissue damage.
Conflict of interest statement
Authors declare no competing financial interests.
Figures
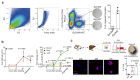
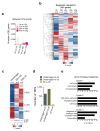

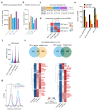

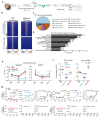




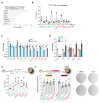
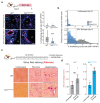
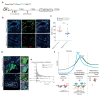
References
Publication types
MeSH terms
Substances
Grants and funding
- A14492/CRUK_/Cancer Research UK/United Kingdom
- 669198/ERC_/European Research Council/International
- 105839/WT_/Wellcome Trust/United Kingdom
- 103792/WT_/Wellcome Trust/United Kingdom
- NC/R001162/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- MR/P016839/1/MRC_/Medical Research Council/United Kingdom
- 104640/WT_/Wellcome Trust/United Kingdom
- MR/K017047/1/MRC_/Medical Research Council/United Kingdom
- A18583/CRUK_/Cancer Research UK/United Kingdom
- 104151/WT_/Wellcome Trust/United Kingdom
- MC_PC_12009/MRC_/Medical Research Council/United Kingdom
- 092096/WT_/Wellcome Trust/United Kingdom
- 18583/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources

