Dietary L-serine confers a competitive fitness advantage to Enterobacteriaceae in the inflamed gut
- PMID: 31686025
- PMCID: PMC6925351
- DOI: 10.1038/s41564-019-0591-6
Dietary L-serine confers a competitive fitness advantage to Enterobacteriaceae in the inflamed gut
Abstract
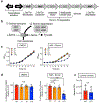
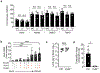
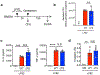
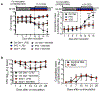
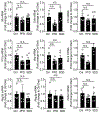
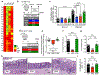
Metabolic reprogramming is associated with the adaptation of host cells to the disease environment, such as inflammation and cancer. However, little is known about microbial metabolic reprogramming or the role it plays in regulating the fitness of commensal and pathogenic bacteria in the gut. Here, we report that intestinal inflammation reprograms the metabolic pathways of Enterobacteriaceae, such as Escherichia coli LF82, in the gut to adapt to the inflammatory environment. We found that E. coli LF82 shifts its metabolism to catabolize L-serine in the inflamed gut in order to maximize its growth potential. However, L-serine catabolism has a minimal effect on its fitness in the healthy gut. In fact, the absence of genes involved in L-serine utilization reduces the competitive fitness of E. coli LF82 and Citrobacter rodentium only during inflammation. The concentration of luminal L-serine is largely dependent on dietary intake. Accordingly, withholding amino acids from the diet markedly reduces their availability in the gut lumen. Hence, inflammation-induced blooms of E. coli LF82 are significantly blunted when amino acids-particularly L-serine-are removed from the diet. Thus, the ability to catabolize L-serine increases bacterial fitness and provides Enterobacteriaceae with a growth advantage against competitors in the inflamed gut.
Conflict of interest statement
Figures







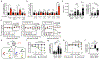
Comment in
-
How to thrive in the inflamed gut.Nat Microbiol. 2020 Jan;5(1):10-11. doi: 10.1038/s41564-019-0642-z. Nat Microbiol. 2020. PMID: 31857729 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

