The ProteomeXchange consortium in 2020: enabling 'big data' approaches in proteomics
- PMID: 31686107
- PMCID: PMC7145525
- DOI: 10.1093/nar/gkz984
The ProteomeXchange consortium in 2020: enabling 'big data' approaches in proteomics
Abstract
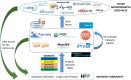
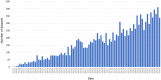
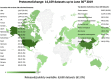
The ProteomeXchange (PX) consortium of proteomics resources (http://www.proteomexchange.org) has standardized data submission and dissemination of mass spectrometry proteomics data worldwide since 2012. In this paper, we describe the main developments since the previous update manuscript was published in Nucleic Acids Research in 2017. Since then, in addition to the four PX existing members at the time (PRIDE, PeptideAtlas including the PASSEL resource, MassIVE and jPOST), two new resources have joined PX: iProX (China) and Panorama Public (USA). We first describe the updated submission guidelines, now expanded to include six members. Next, with current data submission statistics, we demonstrate that the proteomics field is now actively embracing public open data policies. At the end of June 2019, more than 14 100 datasets had been submitted to PX resources since 2012, and from those, more than 9 500 in just the last three years. In parallel, an unprecedented increase of data re-use activities in the field, including 'big data' approaches, is enabling novel research and new data resources. At last, we also outline some of our future plans for the coming years.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S. et al. .. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017; 45:D1100–D1106. - PMC - PubMed
-
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. et al. .. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

