miR-92b-3p Functions As A Key Gene In Esophageal Squamous Cell Cancer As Determined By Co-Expression Analysis
- PMID: 31686859
- PMCID: PMC6799829
- DOI: 10.2147/OTT.S220823
miR-92b-3p Functions As A Key Gene In Esophageal Squamous Cell Cancer As Determined By Co-Expression Analysis
Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is a highly aggressive malignancy. The aims of the present study were to screen the critical miRNA and corresponding target genes that related to development of ESCC by weighted gene correlation network analysis (WGCNA) and investigate the functions by experimental validation.
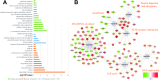
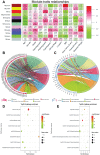
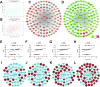
Methods: Datasets of mRNA and miRNA expression data were downloaded from GEO. The R software was used for data preprocessing and differential expression gene analysis. The differentially expressed protein-coding genes (DEGs) and miRNAs (DEMs) were selected (FDR <0.05 or |Fold Change (FC)| >1.5). Meanwhile, 81 expression data of ESCC patients in TCGA combined with clinic information were applied by WGCNA to create networks. The correlational analyses between each module and clinical parameters were conducted, and enrichment analyses of GO and KEGG were subsequently performed. Then, a series of experiments were conducted in ESCC cells by use of miRNA mimics.
Results: In total, 4,023 DEGs and 328 DEMs were screened. After checking good genes and samples, 3,841 genes (3,696 DEGs and 145 DEMs) were used for WGCNA. As a consequence, altogether 11 gene modules were found. Among them, the brown modules were found to be strongly inversely associated with pathological grade. Meanwhile, has-mir-92b, the only miRNA in brown module, had a positive correlation with grade and negatively correlated with potential target gene (KFL4 and DCS2) in the same module. Furthermore, an increased expression of miR-92b-3p and down-regulated KLF4 and DSC2 protein was detected in the ESCC clinical samples. Up-regulated miR-92b-3p shortened G0/G1 phase and promote ESCC cells invasion and migration. Furthermore, we verified that DSC2 and KFL4 was target genes of miR-92b-3p by luciferase report assay.
Conclusion: WGCNA is an efficient approach to system biology. By this procedure, miR-92b-3p was identified as an ESCC-promoting gene by target KLF4 and DCS2.
Keywords: DSC2; ESCC; KLF4; WGCNA; bioinformatics; esophageal squamous cell carcinoma; mir-92b; weighted gene co-expression network analysis.
© 2019 Wang et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

