Association of Genetic Variants With Primary Open-Angle Glaucoma Among Individuals With African Ancestry
- PMID: 31688885
- PMCID: PMC6865235
- DOI: 10.1001/jama.2019.16161
Association of Genetic Variants With Primary Open-Angle Glaucoma Among Individuals With African Ancestry
Abstract
Importance: Primary open-angle glaucoma presents with increased prevalence and a higher degree of clinical severity in populations of African ancestry compared with European or Asian ancestry. Despite this, individuals of African ancestry remain understudied in genomic research for blinding disorders.
Objectives: To perform a genome-wide association study (GWAS) of African ancestry populations and evaluate potential mechanisms of pathogenesis for loci associated with primary open-angle glaucoma.
Design, settings, and participants: A 2-stage GWAS with a discovery data set of 2320 individuals with primary open-angle glaucoma and 2121 control individuals without primary open-angle glaucoma. The validation stage included an additional 6937 affected individuals and 14 917 unaffected individuals using multicenter clinic- and population-based participant recruitment approaches. Study participants were recruited from Ghana, Nigeria, South Africa, the United States, Tanzania, Britain, Cameroon, Saudi Arabia, Brazil, the Democratic Republic of the Congo, Morocco, Peru, and Mali from 2003 to 2018. Individuals with primary open-angle glaucoma had open iridocorneal angles and displayed glaucomatous optic neuropathy with visual field defects. Elevated intraocular pressure was not included in the case definition. Control individuals had no elevated intraocular pressure and no signs of glaucoma.
Exposures: Genetic variants associated with primary open-angle glaucoma.
Main outcomes and measures: Presence of primary open-angle glaucoma. Genome-wide significance was defined as P < 5 × 10-8 in the discovery stage and in the meta-analysis of combined discovery and validation data.
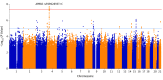
Results: A total of 2320 individuals with primary open-angle glaucoma (mean [interquartile range] age, 64.6 [56-74] years; 1055 [45.5%] women) and 2121 individuals without primary open-angle glaucoma (mean [interquartile range] age, 63.4 [55-71] years; 1025 [48.3%] women) were included in the discovery GWAS. The GWAS discovery meta-analysis demonstrated association of variants at amyloid-β A4 precursor protein-binding family B member 2 (APBB2; chromosome 4, rs59892895T>C) with primary open-angle glaucoma (odds ratio [OR], 1.32 [95% CI, 1.20-1.46]; P = 2 × 10-8). The association was validated in an analysis of an additional 6937 affected individuals and 14 917 unaffected individuals (OR, 1.15 [95% CI, 1.09-1.21]; P < .001). Each copy of the rs59892895*C risk allele was associated with increased risk of primary open-angle glaucoma when all data were included in a meta-analysis (OR, 1.19 [95% CI, 1.14-1.25]; P = 4 × 10-13). The rs59892895*C risk allele was present at appreciable frequency only in African ancestry populations. In contrast, the rs59892895*C risk allele had a frequency of less than 0.1% in individuals of European or Asian ancestry.
Conclusions and relevance: In this genome-wide association study, variants at the APBB2 locus demonstrated differential association with primary open-angle glaucoma by ancestry. If validated in additional populations this finding may have implications for risk assessment and therapeutic strategies.
Conflict of interest statement
Figures


Comment in
-
Genome-Wide Association Studies.JAMA. 2019 Nov 5;322(17):1705-1706. doi: 10.1001/jama.2019.16479. JAMA. 2019. PMID: 31688871 No abstract available.
References
-
- Shiga Y, Akiyama M, Nishiguchi KM, et al. ; Japan Glaucoma Society Omics Group (JGS-OG); NEIGHBORHOOD Consortium . Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum Mol Genet. 2018;27(8):1486-1496. doi:10.1093/hmg/ddy053 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK105556/DK/NIDDK NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- P30 EY014104/EY/NEI NIH HHS/United States
- P30 EY025580/EY/NEI NIH HHS/United States
- R01 EY021818/EY/NEI NIH HHS/United States
- R01 EY020894/EY/NEI NIH HHS/United States
- U54 HG009826/HG/NHGRI NIH HHS/United States
- R01 EY015473/EY/NEI NIH HHS/United States
- R01 DK107786/DK/NIDDK NIH HHS/United States
- T32 GM007337/GM/NIGMS NIH HHS/United States
- R01 DK066358/DK/NIDDK NIH HHS/United States
- R01 DK110113/DK/NIDDK NIH HHS/United States
- P30 EY022589/EY/NEI NIH HHS/United States
- R01 DK087914/DK/NIDDK NIH HHS/United States
- U01 HG004728/HG/NHGRI NIH HHS/United States
- R01 EY023512/EY/NEI NIH HHS/United States
- R01 AG063689/AG/NIA NIH HHS/United States
- R21 EY028671/EY/NEI NIH HHS/United States
- P30 AG028377/AG/NIA NIH HHS/United States
- R01 EY010008/EY/NEI NIH HHS/United States
- R01 DK053591/DK/NIDDK NIH HHS/United States
- R01 EY027004/EY/NEI NIH HHS/United States
- R01 DK070941/DK/NIDDK NIH HHS/United States
- U01 HG007417/HG/NHGRI NIH HHS/United States
- R01 EY019869/EY/NEI NIH HHS/United States
- R01 EY023704/EY/NEI NIH HHS/United States
- R01 EY022305/EY/NEI NIH HHS/United States
- I01 RX001481/RX/RRD VA/United States
- R21 EY029609/EY/NEI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- P30 EY031631/EY/NEI NIH HHS/United States
- R01 DK101855/DK/NIDDK NIH HHS/United States

