The mutational landscape of quinolone resistance in Escherichia coli
- PMID: 31689338
- PMCID: PMC6830822
- DOI: 10.1371/journal.pone.0224650
The mutational landscape of quinolone resistance in Escherichia coli
Abstract
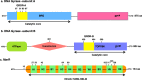
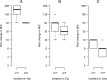
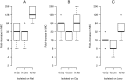
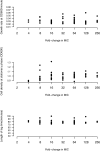
The evolution of antibiotic resistance is influenced by a variety of factors, including the availability of resistance mutations, and the pleiotropic effects of such mutations. Here, we isolate and characterize chromosomal quinolone resistance mutations in E. coli, in order to gain a systematic understanding of the rate and consequences of resistance to this important class of drugs. We isolated over fifty spontaneous resistance mutants on nalidixic acid, ciprofloxacin, and levofloxacin. This set of mutants includes known resistance mutations in gyrA, gyrB, and marR, as well as two novel gyrB mutations. We find that, for most mutations, resistance tends to be higher to nalidixic acid than relative to the other two drugs. Resistance mutations had deleterious impacts on one or more growth parameters, suggesting that quinolone resistance mutations are generally costly. Our findings suggest that the prevalence of specific gyrA alleles amongst clinical isolates are driven by high levels of resistance, at no more cost than other resistance alleles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Public health agency of Canada (PHAC). (2015–2016). Available from: http://www.phac-aspc.gc.ca/rpp/index-eng.php.
-
- Canadian antimicrobial resistance alliance (CARA). CANWARD susceptibility report. 2015. Available from:www.can-r.com.
-
- Gross M. Antibiotics in crisis. Curr Biol. 2013;23(24):1063–1065. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

