Genome Editing in Plants: Exploration of Technological Advancements and Challenges
- PMID: 31689989
- PMCID: PMC6912757
- DOI: 10.3390/cells8111386
Genome Editing in Plants: Exploration of Technological Advancements and Challenges
Abstract
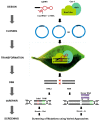
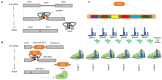
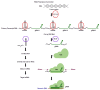
Genome-editing, a recent technological advancement in the field of life sciences, is one of the great examples of techniques used to explore the understanding of the biological phenomenon. Besides having different site-directed nucleases for genome editing over a decade ago, the CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein) based genome editing approach has become a choice of technique due to its simplicity, ease of access, cost, and flexibility. In the present review, several CRISPR/Cas based approaches have been discussed, considering recent advances and challenges to implicate those in the crop improvement programs. Successful examples where CRISPR/Cas approach has been used to improve the biotic and abiotic stress tolerance, and traits related to yield and plant architecture have been discussed. The review highlights the challenges to implement the genome editing in polyploid crop plants like wheat, canola, and sugarcane. Challenges for plants difficult to transform and germline-specific gene expression have been discussed. We have also discussed the notable progress with multi-target editing approaches based on polycistronic tRNA processing, Csy4 endoribonuclease, intron processing, and Drosha ribonuclease. Potential to edit multiple targets simultaneously makes it possible to take up more challenging tasks required to engineer desired crop plants. Similarly, advances like precision gene editing, promoter bashing, and methylome-editing will also be discussed. The present review also provides a catalog of available computational tools and servers facilitating designing of guide-RNA targets, construct designs, and data analysis. The information provided here will be useful for the efficient exploration of technological advances in genome editing field for the crop improvement programs.
Keywords: CRISPR/Cas; biotic and abiotic stress tolerance; methylome-editing; multi-target editing; plant transformation; promoter bashing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
CRISPR/Cas9 for plant genome editing: accomplishments, problems and prospects.Plant Cell Rep. 2016 Jul;35(7):1417-27. doi: 10.1007/s00299-016-1985-z. Epub 2016 Apr 25. Plant Cell Rep. 2016. PMID: 27114166 Review.
-
Current technological interventions and applications of CRISPR/Cas for crop improvement.Mol Biol Rep. 2022 Jun;49(6):5751-5770. doi: 10.1007/s11033-021-06926-5. Epub 2021 Nov 22. Mol Biol Rep. 2022. PMID: 34807378 Review.
-
CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants.Mol Plant. 2017 Mar 6;10(3):530-532. doi: 10.1016/j.molp.2017.01.003. Epub 2017 Jan 13. Mol Plant. 2017. PMID: 28089950 No abstract available.
-
[Genome editing in plants directed by CRISPR/Cas ribonucleoprotein complexes].Yi Chuan. 2020 Jun 20;42(6):556-564. doi: 10.16288/j.yczz.20-017. Yi Chuan. 2020. PMID: 32694114 Review. Chinese.
-
CRISPR/Cas: A powerful tool for gene function study and crop improvement.J Adv Res. 2020 Oct 21;29:207-221. doi: 10.1016/j.jare.2020.10.003. eCollection 2021 Mar. J Adv Res. 2020. PMID: 33842017 Free PMC article. Review.
Cited by
-
Plant Polyphenols-Biofortified Foods as a Novel Tool for the Prevention of Human Gut Diseases.Antioxidants (Basel). 2020 Dec 3;9(12):1225. doi: 10.3390/antiox9121225. Antioxidants (Basel). 2020. PMID: 33287404 Free PMC article. Review.
-
A Universal System of CRISPR/Cas9-Mediated Gene Targeting Using All-in-One Vector in Plants.Front Genome Ed. 2020 Nov 25;2:604289. doi: 10.3389/fgeed.2020.604289. eCollection 2020. Front Genome Ed. 2020. PMID: 34713227 Free PMC article.
-
CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis in Sesame (Sesamum indicum L.).Front Plant Sci. 2022 Jul 11;13:935825. doi: 10.3389/fpls.2022.935825. eCollection 2022. Front Plant Sci. 2022. PMID: 35898225 Free PMC article.
-
The applications of CRISPR/Cas-mediated microRNA and lncRNA editing in plant biology: shaping the future of plant non-coding RNA research.Planta. 2023 Dec 28;259(2):32. doi: 10.1007/s00425-023-04303-z. Planta. 2023. PMID: 38153530 Review.
-
Integration of CRISPR/Cas9 with multi-omics technologies to engineer secondary metabolite productions in medicinal plant: Challenges and Prospects.Funct Integr Genomics. 2024 Nov 4;24(6):207. doi: 10.1007/s10142-024-01486-w. Funct Integr Genomics. 2024. PMID: 39496976 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

