Functional diversification of the chemical landscapes of yeast Sec14-like phosphatidylinositol transfer protein lipid-binding cavities
- PMID: 31690622
- PMCID: PMC6916498
- DOI: 10.1074/jbc.RA119.011153
Functional diversification of the chemical landscapes of yeast Sec14-like phosphatidylinositol transfer protein lipid-binding cavities
Erratum in
-
Correction: Functional diversification of the chemical landscapes of yeast Sec14-like phosphatidylinositol transfer protein lipid-binding cavities.J Biol Chem. 2020 Jan 31;295(5):1368. doi: 10.1074/jbc.AAC120.012555. J Biol Chem. 2020. PMID: 32005645 Free PMC article. No abstract available.
Abstract
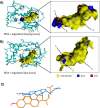
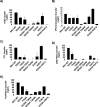
Phosphatidylinositol-transfer proteins (PITPs) are key regulators of lipid signaling in eukaryotic cells. These proteins both potentiate the activities of phosphatidylinositol (PtdIns) 4-OH kinases and help channel production of specific pools of phosphatidylinositol 4-phosphate (PtdIns(4)P) dedicated to specific biological outcomes. In this manner, PITPs represent a major contributor to the mechanisms by which the biological outcomes of phosphoinositide are diversified. The two-ligand priming model proposes that the engine by which Sec14-like PITPs potentiate PtdIns kinase activities is a heterotypic lipid-exchange cycle where PtdIns is a common exchange substrate among the Sec14-like PITP family, but the second exchange ligand varies with the PITP. A major prediction of this model is that second-exchangeable ligand identity will vary from PITP to PITP. To address the heterogeneity in the second exchange ligand for Sec14-like PITPs, we used structural, computational, and biochemical approaches to probe the diversities of the lipid-binding cavity microenvironments of the yeast Sec14-like PITPs. The collective data report that yeast Sec14-like PITP lipid-binding pockets indeed define diverse chemical microenvironments that translate into differential ligand-binding specificities across this protein family.
Keywords: Sec14-domain; cavity mapping; computational biology; lipid metabolism; phosphatidylinositol transfer proteins; phosphoinositide; signaling; squalene; sterol.
© 2019 Tripathi et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Schaaf G., Ortlund E. A., Tyeryar K. R., Mousley C. J., Ile K. E., Garrett T. A., Ren J., Woolls M. J., Raetz C. R., Redinbo M. R., and Bankaitis V. A. (2008) The functional anatomy of PL binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily. Mol. Cell 29, 191–206 10.1016/j.molcel.2007.11.026 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

