Sex- and tissue-specific transcriptome analyses and expression profiling of olfactory-related genes in Ceracris nigricornis Walker (Orthoptera: Acrididae)
- PMID: 31694535
- PMCID: PMC6836668
- DOI: 10.1186/s12864-019-6208-x
Sex- and tissue-specific transcriptome analyses and expression profiling of olfactory-related genes in Ceracris nigricornis Walker (Orthoptera: Acrididae)
Abstract
Background: The sophisticated insect olfactory system plays an important role in recognizing external odors and enabling insects to adapt to environment. Foraging, host seeking, mating, ovipositing and other forms of chemical communication are based on olfaction, which requires the participation of multiple olfactory genes. The exclusive evolutionary trend of the olfactory system in Orthoptera insects is an excellent model for studying olfactory evolution, but limited olfaction research is available for these species. The olfactory-related genes of Ceracris nigricornis Walker (Orthoptera: Acrididae), a severe pest of bamboos, have not yet been reported.
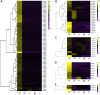
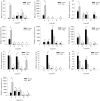
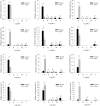
Results: We sequenced and analyzed the transcriptomes from different tissues of C. nigricornis and obtained 223.76 Gb clean data that were assembled into 43,603 unigenes with an N50 length of 2235 bp. Among the transcripts, 66.79% of unigenes were annotated. Based on annotation and tBLASTn results, 112 candidate olfactory-related genes were identified for the first time, including 20 odorant-binding proteins (OBPs), 10 chemosensory-binding proteins (CSPs), 71 odorant receptors (ORs), eight ionotropic receptors (IRs) and three sensory neuron membrane proteins (SNMPs). The fragments per kilobase per million mapped fragments (FPKM) values showed that most olfactory-related differentially expressed genes (DEGs) were enriched in the antennae, and these results were confirmed by detecting the expression of olfactory-related genes with quantitative real-time PCR (qRT-PCR). Among these antennae-enriched genes, some were sex-biased, indicating their different roles in the olfactory system of C. nigricornis.
Conclusions: This study provides the first comprehensive list and expression profiles of olfactory-related genes in C. nigricornis and a foundation for functional studies of these olfactory-related genes at the molecular level.
Keywords: Ceracris nigricornis; Chemosensory-binding protein; Expression profiles analysis; Ionotropic receptor; Odorant receptor; Odorant-binding protein; Sensory neuron membrane protein; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Bian L, Sun X, Luo Z, Zhang Z, Chen Z. Design and selection of trap color for capture of the tea leafhopper, Empoasca vitis, by orthogonal optimization. Entomologia Experimentalis Et Applicata. 2014;151(3):247–258. doi: 10.1111/eea.12191. - DOI
-
- Jin S, Chen ZM, Backus EA, Sun XL, Xiao B. Characterization of EPG waveforms for the tea green leafhopper, Empoasca vitis Göthe (Hemiptera: Cicadellidae), on tea plants and their correlation with stylet activities. J Insect Physiol. 2012;58(9):1235–1244. doi: 10.1016/j.jinsphys.2012.06.008. - DOI - PubMed
-
- Song T, Wang K, Peng W, Wang J, Xiao R, Zeng F, Tang Y. Ecological effects of intercropping white clover on tea plantation in a subtropical hilly region. Acta Ecol Sin. 2006;26(11):3647–3655.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

