Reduced Fitness Costs of mcr-1.2 Compared to Mutated pmrB in Isogenic Colistin-Resistant KPC-3-Producing Klebsiella pneumoniae
- PMID: 31694895
- PMCID: PMC6835208
- DOI: 10.1128/mSphere.00551-19
Reduced Fitness Costs of mcr-1.2 Compared to Mutated pmrB in Isogenic Colistin-Resistant KPC-3-Producing Klebsiella pneumoniae
Abstract
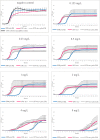
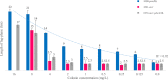
In the present study, we provide the results of a detailed genomic analysis and the growth characteristics of a colistin-resistant KPC-3-producing Klebsiella pneumoniae sequence type 512 (ST512) isolate (the colR-KPC3-KP isolate) with a mutated pmrB and isogenic isolates of colR-KPC3-KP with mcr-1.2 isolated from an immunocompromised patient. From 2014 to 2017, four colR-KPC3-KP isolates were detected in rectal swab samples collected from a pediatric hematology patient at the Azienda Ospedaliero-Universitaria Pisana in Pisa, Italy. Whole-genome sequencing was performed by MiSeq sequencing (Illumina). Growth experiments were performed using different concentrations of colistin. The growth lag phases both of an isolate harboring a deletion in pmrB and of clonal variants with mcr-1.2 were assessed by the use of real-time light-scattering measurements. In the first isolate (isolate 1000-pmrBΔ, recovered in September 2014), a 17-nucleotide deletion in pmrB was detected. In subsequent isolates, the mcr-1.2 gene associated with the plasmid pIncX4-AOUP was found, while pmrB was intact. Additionally, plasmid pIncQ-AOUP, harboring aminoglycoside resistance genes, was detected. The growth curves of the first three isolates were identical without colistin exposure; however, at higher concentrations of colistin, the growth curves of the isolate with a deletion in pmrB showed longer lag phases. We observed the replacement of mutated colR-KPC3-KP pmrB by isogenic isolates with multiple resistance plasmids, including mcr-1.2-carrying pIncX4, probably due to coselection under gentamicin treatment in a patient with prolonged colR-KPC3-KP carriage. The carriage of these isolates persisted in follow-up cultures. Coselection and the advantages in growth characteristics suggest that the plasmid-mediated resistance conferred by mcr has fewer fitness costs in colR-KPC3-KP than mutations in chromosomal pmrB, contributing to the success of this highly resistant hospital-adapted epidemiological lineage.IMPORTANCE Our study shows a successful prolonged human colonization by a colistin-resistant Klebsiella pneumoniae isolate harboring mcr-1.2 An intense antibiotic therapy contributed to the maintenance of this microorganism through the acquisition of new resistance genes. The isolates carrying mcr-1.2 showed fewer fitness costs than isogenic isolates with a pmrB mutation in the chromosome. Coselection and reduced fitness costs may explain the replacement of isolates with the pmrB mutation by other isolates and the ability of the microorganism to persist despite antibiotic treatment.
Keywords: colistin; fitness cost; mcr-1.2; molecular epidemiology.
Copyright © 2019 Giordano et al.
Figures

References
-
- Giordano C, Barnini S, Tsioutis C, Chlebowicz MA, Scoulica EV, Gikas A, Rossen JW, Friedrich AW, Bathoorn E. 2018. Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: the role of ISL3 on plasmids. Int J Antimicrob Agents 51:260–265. doi:10.1016/j.ijantimicag.2017.10.011. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

