E2FB Interacts with RETINOBLASTOMA RELATED and Regulates Cell Proliferation during Leaf Development
- PMID: 31694902
- PMCID: PMC6945829
- DOI: 10.1104/pp.19.00212
E2FB Interacts with RETINOBLASTOMA RELATED and Regulates Cell Proliferation during Leaf Development
Abstract
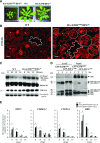
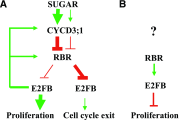
Cell cycle entry and quiescence are regulated by the E2F transcription factors in association with RETINOBLASTOMA-RELATED (RBR). E2FB is considered to be a transcriptional activator of cell cycle genes, but its function during development remains poorly understood. Here, by studying E2FB-RBR interaction, E2F target gene expression, and epidermal cell number and shape in e2fb mutant and overexpression lines during leaf development in Arabidopsis (Arabidopsis thaliana), we show that E2FB in association with RBR plays a role in the inhibition of cell proliferation to establish quiescence. In young leaves, both RBR and E2FB are abundant and form a repressor complex that is reinforced by an autoregulatory loop. Increased E2FB levels, either by expression driven by its own promoter or ectopically together with DIMERIZATION PARTNER A, further elevate the amount of this repressor complex, leading to reduced leaf cell number. Cell overproliferation in e2fb mutants and in plants overexpressing a truncated form of E2FB lacking the RBR binding domain strongly suggested that RBR repression specifically acts through E2FB. The increased number of small cells below the guard cells and of fully developed stomata indicated that meristemoids preferentially hyperproliferate. As leaf development progresses and cells differentiate, the amount of RBR and E2FB gradually declined. At this stage, elevation of E2FB level can overcome RBR repression, leading to reactivation of cell division in pavement cells. In summary, E2FB in association with RBR is central to regulating cell proliferation during organ development to determine final leaf cell number.
© 2020 The authors. All Rights Reserved.
Figures
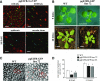
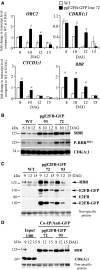


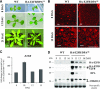
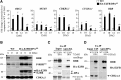
Similar articles
-
Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes.EMBO J. 2012 Mar 21;31(6):1480-93. doi: 10.1038/emboj.2012.13. Epub 2012 Feb 3. EMBO J. 2012. PMID: 22307083 Free PMC article.
-
Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development.Plant Physiol. 2006 Jan;140(1):67-80. doi: 10.1104/pp.105.071027. Epub 2005 Dec 16. Plant Physiol. 2006. PMID: 16361519 Free PMC article.
-
S6K1 and E2FB are in mutually antagonistic regulatory links controlling cell growth and proliferation in Arabidopsis.Plant Signal Behav. 2013 Jun;8(6):e24367. doi: 10.4161/psb.24367. Epub 2013 Mar 26. Plant Signal Behav. 2013. PMID: 23531690 Free PMC article.
-
Molecular networks regulating cell division during Arabidopsis leaf growth.J Exp Bot. 2020 Apr 23;71(8):2365-2378. doi: 10.1093/jxb/erz522. J Exp Bot. 2020. PMID: 31748815 Free PMC article. Review.
-
Leaf growth - complex regulation of a seemingly simple process.Plant J. 2024 Feb;117(4):1018-1051. doi: 10.1111/tpj.16558. Epub 2023 Nov 27. Plant J. 2024. PMID: 38012838 Review.
Cited by
-
The canonical E2Fs together with RETINOBLASTOMA-RELATED are required to establish quiescence during plant development.Commun Biol. 2023 Sep 4;6(1):903. doi: 10.1038/s42003-023-05259-2. Commun Biol. 2023. PMID: 37666980 Free PMC article.
-
The genomic basis of the plant island syndrome in Darwin's giant daisies.Nat Commun. 2022 Jun 28;13(1):3729. doi: 10.1038/s41467-022-31280-w. Nat Commun. 2022. PMID: 35764640 Free PMC article.
-
Genome-wide identification and functional characterization of the PheE2F/DP gene family in Moso bamboo.BMC Plant Biol. 2021 Mar 29;21(1):158. doi: 10.1186/s12870-021-02924-8. BMC Plant Biol. 2021. PMID: 33781213 Free PMC article.
-
The Arabidopsis GRAS-type SCL28 transcription factor controls the mitotic cell cycle and division plane orientation.Proc Natl Acad Sci U S A. 2021 Feb 9;118(6):e2005256118. doi: 10.1073/pnas.2005256118. Proc Natl Acad Sci U S A. 2021. PMID: 33526654 Free PMC article.
-
Molecular Analysis of the E2F/DP Gene Family of Daucus carota and Involvement of the DcE2F1 Factor in Cell Proliferation.Front Plant Sci. 2021 Mar 12;12:652570. doi: 10.3389/fpls.2021.652570. eCollection 2021. Front Plant Sci. 2021. PMID: 33777085 Free PMC article.
References
-
- Abraham Z, del Pozo JC (2012) Ectopic expression of E2FB, a cell cycle transcription factor, accelerates flowering and increases fruit yield in tomato. J Plant Growth Regul 31: 11–24
-
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: A not-so-gradual process. Dev Cell 22: 64–78 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

