Intact Viral Particle Counts Measured by Flow Virometry Provide Insight into the Infectivity and Genome Packaging Efficiency of Moloney Murine Leukemia Virus
- PMID: 31694951
- PMCID: PMC6955258
- DOI: 10.1128/JVI.01600-19
Intact Viral Particle Counts Measured by Flow Virometry Provide Insight into the Infectivity and Genome Packaging Efficiency of Moloney Murine Leukemia Virus
Abstract
Murine leukemia viruses (MLVs) have long been used as a research model to further our understanding of retroviruses. These simple gammaretroviruses have been studied extensively in various facets of science for nearly half a century, yet we have surprisingly little quantitative information about some of the basic features of these viral particles. These include parameters such as the genome packaging efficiency and the number of particles required for a productive infection. The reason for this knowledge gap relies primarily on the technical challenge of accurately measuring intact viral particles from infected cell supernatants. Virus-infected cells are well known to release soluble viral proteins, defective viruses, and extracellular vesicles (EVs) harboring viral proteins that may mimic viruses, all of which can skew virus titer quantifications. Flow virometry, also known as nanoscale flow cytometry or simply small-particle flow cytometry, is an emerging analytical method enabling high-throughput single-virus phenotypic characterizations. By utilizing the viral envelope glycoprotein (Env) and monodisperse light scattering characteristics as discerning parameters of intact virus particles, here, we analyzed the basic properties of Moloney MLV (M-MLV). We show that <24% of the total p30 capsid protein measured in infected cell supernatants is associated with intact viruses. We calculate that about one in five M-MLV particles contains a viral RNA genome pair and that individual intact particle infectivity is about 0.4%. These findings provide new insights into the characteristics of an extensively studied prototypical retrovirus while highlighting the benefits of flow virometry for the field of virology.IMPORTANCE Gammaretroviruses, or, more specifically, murine leukemia viruses (MLVs), have been a longstanding model for studying retroviruses. Although being extensively analyzed and dissected for decades, several facets of MLV biology are still poorly understood. One of the primary challenges has been enumerating total intact virus particles in a sample. While several analytical methods can precisely measure virus protein amounts, MLVs are known to induce the secretion of soluble and vesicle-associated viral proteins that can skew these measurements. With recent technological advances in flow cytometry, it is now possible to analyze viruses down to 90 nm in diameter with an approach called flow virometry. The technique has the added benefit of being able to discriminate viruses from extracellular vesicles and free viral proteins in order to confidently provide an intact viral particle count. Here, we used flow virometry to provide new insights into the basic characteristics of Moloney MLV.
Keywords: flow virometry; genome packaging; intact particle count; murine leukemia virus.
Copyright © 2020 American Society for Microbiology.
Figures
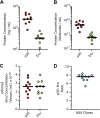




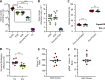
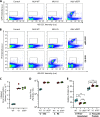
References
-
- Moloney JB. 1960. Biological studies on a lymphoid-leukemia virus extracted from sarcoma 37. I. Origin and introductory investigations. J Natl Cancer Inst 24:933–951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

