UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor
- PMID: 31695187
- PMCID: PMC6858872
- DOI: 10.1038/s41594-019-0324-9
UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor
Abstract
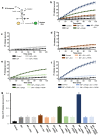
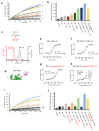
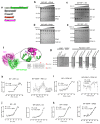
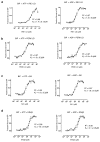
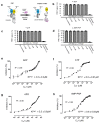
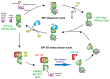
BiP is a major endoplasmic reticulum (ER) chaperone and is suggested to act as primary sensor in the activation of the unfolded protein response (UPR). How BiP operates as a molecular chaperone and as an ER stress sensor is unknown. Here, by reconstituting components of human UPR, ER stress and BiP chaperone systems, we discover that the interaction of BiP with the luminal domains of UPR proteins IRE1 and PERK switch BiP from its chaperone cycle into an ER stress sensor cycle by preventing the binding of its co-chaperones, with loss of ATPase stimulation. Furthermore, misfolded protein-dependent dissociation of BiP from IRE1 is primed by ATP but not ADP. Our data elucidate a previously unidentified mechanistic cycle of BiP function that explains its ability to act as an Hsp70 chaperone and ER stress sensor.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Hetz C, Papa FR. The Unfolded Protein Response and Cell Fate Control. Mol Cell. 2017 - PubMed
-
- Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. - PubMed
-
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

