Bread Wheat With High Salinity and Sodicity Tolerance
- PMID: 31695711
- PMCID: PMC6817574
- DOI: 10.3389/fpls.2019.01280
Bread Wheat With High Salinity and Sodicity Tolerance
Abstract
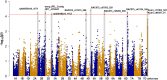
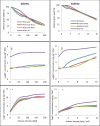
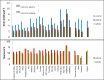
Soil salinity and sodicity are major constraints to global cereal production, but breeding for tolerance has been slow. Narrow gene pools, over-emphasis on the sodium (Na+) exclusion mechanism, little attention to osmotic stress/tissue tolerance mechanism(s) in which accumulation of inorganic ions such as Na+ is implicated, and lack of a suitable screening method have impaired progress. The aims of this study were to discover novel genes for Na+ accumulation using genome-wide association studies, compare growth responses to salinity and sodicity in low-Na+ bread Westonia with Nax1 and Nax2 genes and high-Na+ bread wheat Baart-46, and evaluate growth responses to salinity and sodicity in bread wheats with varying leaf Na+ concentrations. The novel high-Na+ bread wheat germplasm, MW#293, had higher grain yield under salinity and sodicity, in absolute and relative terms, than the other bread wheat entries tested. Genes associated with high Na+ accumulation in bread wheat were identified, which may be involved in tissue tolerance/osmotic adjustment. As most modern bread wheats are efficient at excluding Na+, further reduction in plant Na+ is unlikely to provide agronomic benefit. The salinity and sodicity tolerant germplasm MW#293 provides an opportunity for the development of future salinity/sodicity tolerant bread wheat.
Keywords: chloride; ionic; osmotic; salinity; sodicity; sodium; sodium humate; tolerance.
Copyright © 2019 Genc, Taylor, Lyons, Li, Cheong, Appelbee, Oldach and Sutton.
Figures




References
-
- Acosta-Motos J. R., Ortuna M. F., Bernal-Vicente A., Diaz-Vivancos P., Sanches-Blanco M. J., Hernandez J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy 7, 18. 10.3390/agronomy7010018 - DOI
-
- Adem G. A., Roy S. R., Huang Y., Chen Z.-H., Wang F., Zhou M., et al. (2017). Expressing Arabidopsis thaliana V-ATPase subunit C in barley (Hordeum vulgare L.) improves plant performance under saline condition by enabling better osmotic adjustment. Funct. Plant Biol. 44, 1147–1159. 10.1071/FP17133 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

