Chlamydia trachomatis: when the virulence-associated genome backbone imports a prevalence-associated major antigen signature
- PMID: 31697227
- PMCID: PMC6927300
- DOI: 10.1099/mgen.0.000313
Chlamydia trachomatis: when the virulence-associated genome backbone imports a prevalence-associated major antigen signature
Abstract
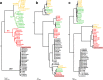
Chlamydia trachomatis is the most prevalent sexually transmitted bacterium worldwide and the causative agent of trachoma. Its strains are classified according to their ompA genotypes, which are strongly linked to differential tissue tropism and disease outcomes [ocular disease, urogenital disease and lymphogranuloma venereum (LGV)]. While the genome-based species phylogenetic tree presents four main clades correlating with tropism/prevalence, namely ocular, LGV, urogenital T1 (more prevalent genotypes) and urogenital T2 (less prevalent genotypes), inter-clade exchange of ompA is considered a rare phenomenon probably mediating marked tropism alterations. An LGV epidemic, associated with the clonal expansion of the L2b genotype, has emerged in the last few decades, raising concerns particularly due to its atypical clinical presentation (ulcerative proctitis) and circulation among men who have sex with men (MSM). Here, we report an LGV outbreak, mostly affecting human immunodeficiency virus-positive MSM engaging in high-risk sexual practices, caused by an L2b strain with a rather unique non-LGV ompA signature that precluded the laboratory notification of this outbreak as LGV. C. trachomatis whole-genome capture and sequencing directly from clinical samples was applied to deeply characterize the genomic backbone of this novel LGV outbreak-causing clone. It revealed a chimeric genome structure due to the genetic transfer of ompA and four neighbouring genes from a serovar D/Da strain, likely possessing the genomic backbone associated with the more prevalent urogenital genotypes (T1 clade), to an LGV (L2b) strain. The hybrid L2b/D-Da strain presents the adhesin and immunodominant antigen MOMP (major outer membrane protein) (encoded by ompA) with an epitope repertoire typical of non-invasive genital strains, while keeping the genome-dispersed virulence fingerprint of a classical LGV strain. As previously reported for inter-clade ompA exchange among non-LGV clades, this novel C. trachomatis genomic mosaic involving a contemporary epidemiologically and clinically relevant LGV strain may have implications on its transmission, tissue tropism and pathogenic capabilities. The emergence of variants with epidemic and pathogenic potential highlights the need for more focused surveillance strategies to capture C. trachomatis evolution in action.
Keywords: Chlamydia trachomatis; inter-clade exchange; lymphogranuloma venereum (LGV); ompA; outbreak; recombination.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Comment in
-
Toward the Spread of the New L2b/D-Da Hybrid Chlamydia trachomatis Strain in Men Who Have Sex With Men in France?Clin Infect Dis. 2021 Sep 15;73(6):1130-1131. doi: 10.1093/cid/ciab253. Clin Infect Dis. 2021. PMID: 33744938 No abstract available.
References
-
- World Health Organization Geneva: World Health Organization; 2018. Report on Global Sexually Transmitted Infection Surveillance.
-
- Nieuwenhuis RF, Ossewaarde JM, Götz HM, Dees J, Thio HB, et al. Resurgence of lymphogranuloma venereum in Western Europe: an outbreak of Chlamydia trachomatis serovar L2 proctitis in the Netherlands among men who have sex with men. Clin Infect Dis. 2004;39:996–1003. doi: 10.1086/423966. - DOI - PubMed
-
- Savage EJ, van de Laar MJ, Gallay A, van der Sande M, Hamouda O, et al. Lymphogranuloma venereum in Europe, 2003-2008. Euro Surveill. 2009;14:19428. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

