Oligomerization of RIG-I and MDA5 2CARD domains
- PMID: 31697400
- PMCID: PMC6954692
- DOI: 10.1002/pro.3776
Oligomerization of RIG-I and MDA5 2CARD domains
Abstract
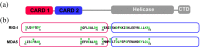
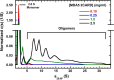
The innate immune system is the first line of defense against invading pathogens. The retinoic acid-inducible gene I (RIG-I) like receptors (RLRs), RIG-I and melanoma differentiation-associated protein 5 (MDA5), are critical for host recognition of viral RNAs. These receptors contain a pair of N-terminal tandem caspase activation and recruitment domains (2CARD), an SF2 helicase core domain, and a C-terminal regulatory domain. Upon RLR activation, 2CARD associates with the CARD domain of MAVS, leading to the oligomerization of MAVS, downstream signaling and interferon induction. Unanchored K63-linked polyubiquitin chains (polyUb) interacts with the 2CARD domain, and in the case of RIG-I, induce tetramer formation. However, the nature of the MDA5 2CARD signaling complex is not known. We have used sedimentation velocity analytical ultracentrifugation to compare MDA5 2CARD and RIG-I 2CARD binding to polyUb and to characterize the assembly of MDA5 2CARD oligomers in the absence of polyUb. Multi-signal sedimentation velocity analysis indicates that Ub4 binds to RIG-I 2CARD with a 3:4 stoichiometry and cooperatively induces formation of an RIG-I 2CARD tetramer. In contrast, Ub4 and Ub7 interact with MDA5 2CARD weakly and form complexes with 1:1 and 2:1 stoichiometries but do not induce 2CARD oligomerization. In the absence of polyUb, MDA5 2CARD self-associates to forms large oligomers in a concentration-dependent manner. Thus, RIG-I and MDA5 2CARD assembly processes are distinct. MDA5 2CARD concentration-dependent self-association, rather than polyUb binding, drives oligomerization and MDA5 2CARD forms oligomers larger than tetramer. We propose a mechanism where MDA5 2CARD oligomers, rather than a stable tetramer, function to nucleate MAVS polymerization.
Keywords: K63-linked polyubiquitin; RIG-I-like receptors; analytical ultracentrifugation; innate immunity; multi-signal sedimentation velocity.
© 2019 The Protein Society.
Figures


References
-
- Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG‐I has an essential function in double‐stranded RNA‐induced innate antiviral responses. Nat Immunol. 2004;5:730–737. - PubMed
-
- Hornung V, Ellegast J, Kim S, et al. 5′‐triphosphate RNA is the ligand for RIG‐I. Science. 2006;314:994–997. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

