Evaluating rectal swab collection method for gut microbiome analysis in the common marmoset (Callithrix jacchus)
- PMID: 31697779
- PMCID: PMC6837755
- DOI: 10.1371/journal.pone.0224950
Evaluating rectal swab collection method for gut microbiome analysis in the common marmoset (Callithrix jacchus)
Abstract
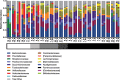
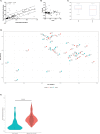
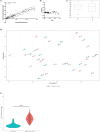
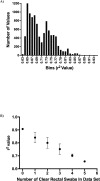
The common marmoset (Callithrix jacchus) is increasingly used as an animal model for biomedical research; however, gastrointestinal diseases causing significant morbidity are endemic in many captive marmoset colonies. Establishing gut microbiome patterns in a marmoset colony may aid in clinical decision-making and model reproducibility. A standardized method of sample collection and storage is essential for proper interpretation of microbiome data. While microbiome studies commonly utilize fecal samples, the goal of this study was to determine whether the microbiome profile from a rectal swab performed on a sedated animal was comparable to the microbiome profile from a fecal sample. During routine physical exams, paired fecal and rectal swab samples were collected from each of 23 marmosets. DNA was extracted from all fecal and rectal swab samples and 16S ribosomal RNA gene sequences were amplified and analyzed. Initial comparison of the relative abundance of bacterial phyla between paired samples had a r2 value of 0.70 with S of 0.08 with no significant differences in α and β diversity metrics between fecal and rectal samples. Initial analysis however, revealed 5 discordant fecal-rectal pairs which corresponded only with the 5 rectal swabs that were classified as free of visible fecal matter during collection. Exclusion of these 5 pairs resulted in an optimized fit of the data as evidenced by a r2 value of 0.91 with S of 0.05. These results demonstrate that rectal swabs are a reliable method for profiling the fecal microbiome in the marmoset since the bacterial composition from a rectal swab with visible fecal contents correlated well with the bacterial composition from a fecal sample from the same marmoset. This study highlights the importance of standardized sample collection methods and exclusion of inappropriate samples.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Marini RP, Wachtman LM, Tardif SD, Mansfield K, Fox JG. The Common Marmoset in Captivity and Biomedical Research. Academic Press; 2018.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

