The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative
- PMID: 31697809
- PMCID: PMC7218752
- DOI: 10.1182/blood.2019002206
The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative
Abstract
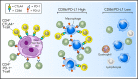
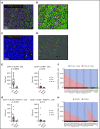
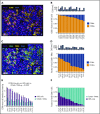
Classic Hodgkin lymphoma (cHL) is a tumor composed of rare, atypical, germinal center-derived B cells (Hodgkin Reed-Sternberg [HRS] cells) embedded within a robust but ineffective inflammatory milieu. The cHL tumor microenvironment (TME) is compartmentalized into "niches" rich in programmed cell death-1 ligand (PD-L1)-positive HRS cells and tumor-associated macrophages (TAMs), which associate with PD-1-positive T cells to suppress antitumor immunity via PD-L1/PD-1 signaling. Despite the exquisite sensitivity of cHL to PD-1 checkpoint blockade, most patients eventually relapse and need therapeutic alternatives. Using multiplex immunofluorescence microscopy with digital image analysis, we found that cHL is highly enriched for non-T-regulatory, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)-positive T cells (compared with reactive lymphoid tissues) that outnumber PD-1-positive and lymphocyte-activating gene-3 (LAG-3)-positive T cells. In addition, T cells touching HRS cells are more frequently positive for CTLA-4 than for PD-1 or LAG-3. We further found that HRS cells, and a subset of TAMs, are positive for the CTLA-4 ligand CD86 and that the fractions of T cells and TAMs that are CTLA-4-positive and CD86-positive, respectively, are greater within a 75 μm HRS cell niche relative to areas outside this region (CTLA-4, 38% vs 18% [P = .0001]; CD86, 38% vs 24% [P = .0007]). Importantly, CTLA-4-positive cells are present, and focally contact HRS cells, in recurrent cHL tumors following a variety of therapies, including PD-1 blockade. These results implicate CTLA-4:CD86 interactions as a component of the immunologically privileged niche surrounding HRS cells and raise the possibility that patients with cHL refractory to PD-1 blockade may benefit from CTLA-4 blockade.
© 2019 by The American Society of Hematology.
Figures





References
-
- Swerdlow SH, Campo E, Harris NL, et al. . WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th Edition Lyon, France: IARC; 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

