Structure of the human metapneumovirus polymerase phosphoprotein complex
- PMID: 31698413
- PMCID: PMC6949429
- DOI: 10.1038/s41586-019-1759-1
Structure of the human metapneumovirus polymerase phosphoprotein complex
Abstract
Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) cause severe respiratory diseases in infants and elderly adults1. No vaccine or effective antiviral therapy currently exists to control RSV or HMPV infections. During viral genome replication and transcription, the tetrameric phosphoprotein P serves as a crucial adaptor between the ribonucleoprotein template and the L protein, which has RNA-dependent RNA polymerase (RdRp), GDP polyribonucleotidyltransferase and cap-specific methyltransferase activities2,3. How P interacts with L and mediates the association with the free form of N and with the ribonucleoprotein is not clear for HMPV or other major human pathogens, including the viruses that cause measles, Ebola and rabies. Here we report a cryo-electron microscopy reconstruction that shows the ring-shaped structure of the polymerase and capping domains of HMPV-L bound to a tetramer of P. The connector and methyltransferase domains of L are mobile with respect to the core. The putative priming loop that is important for the initiation of RNA synthesis is fully retracted, which leaves space in the active-site cavity for RNA elongation. P interacts extensively with the N-terminal region of L, burying more than 4,016 Å2 of the molecular surface area in the interface. Two of the four helices that form the coiled-coil tetramerization domain of P, and long C-terminal extensions projecting from these two helices, wrap around the L protein in a manner similar to tentacles. The structural versatility of the four P protomers-which are largely disordered in their free state-demonstrates an example of a 'folding-upon-partner-binding' mechanism for carrying out P adaptor functions. The structure shows that P has the potential to modulate multiple functions of L and these results should accelerate the design of specific antiviral drugs.
Conflict of interest statement
Figures







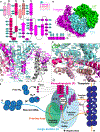
References
-
- Pflug A, Guilligay D, Reich S, Cusack S Structure of Influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–60 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

