Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology
- PMID: 31699565
- PMCID: PMC7020649
- DOI: 10.1016/j.tem.2019.09.007
Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology
Abstract
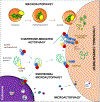
Autophagy contributes to cellular quality control and energetics through lysosomal breakdown and recycling of essential cellular components. Chaperone-mediated autophagy (CMA) adds to these autophagic functions the ability to timely and selectively degrade single tagged proteins to terminate their cellular function and, in this way, participate in the regulation of multiple cellular processes. Many cancer cells upregulate CMA for protumorigenic and prosurvival purposes. However, growing evidence supports a physiological role for CMA in limiting malignant transformation. Understanding the mechanisms behind this functional switch of CMA from antioncogenic to pro-oncogenic is fundamental for targeting CMA in cancer treatment. We summarize current understanding of CMA functions in cancer biology and discuss the basis for its context-dependent dual role in oncogenesis.
Keywords: chaperones; lysosomes; metabolism; oncogenes; protein degradation; tumorigenesis.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures


References
-
- Stolz A. et al. (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16 (6), 495–501. - PubMed
References unique to Boxes
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

