Trisomy 21 dysregulates T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity
- PMID: 31699819
- PMCID: PMC6883781
- DOI: 10.1073/pnas.1908129116
Trisomy 21 dysregulates T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity
Abstract
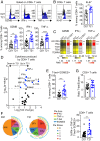
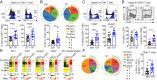
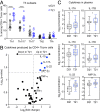
Trisomy 21 (T21) causes Down syndrome (DS), a condition characterized by high prevalence of autoimmune disorders. However, the molecular and cellular mechanisms driving this phenotype remain unclear. Building upon our previous finding that T cells from people with DS show increased expression of interferon (IFN)-stimulated genes, we have completed a comprehensive characterization of the peripheral T cell compartment in adults with DS with and without autoimmune conditions. CD8+ T cells from adults with DS are depleted of naïve subsets and enriched for differentiated subsets, express higher levels of markers of activation and senescence (e.g., IFN-γ, Granzyme B, PD-1, KLRG1), and overproduce cytokines tied to autoimmunity (e.g., TNF-α). Conventional CD4+ T cells display increased differentiation, polarization toward the Th1 and Th1/17 states, and overproduction of the autoimmunity-related cytokines IL-17A and IL-22. Plasma cytokine analysis confirms elevation of multiple autoimmunity-related cytokines (e.g., TNF-α, IL17A-D, IL-22) in people with DS, independent of diagnosis of autoimmunity. Although Tregs are more abundant in DS, functional assays show that CD8+ and CD4+ effector T cells with T21 are resistant to Treg-mediated suppression, regardless of Treg karyotype. Transcriptome analysis of white blood cells and T cells reveals strong signatures of T cell differentiation and activation that correlate positively with IFN hyperactivity. Finally, mass cytometry analysis of 8 IFN-inducible phosphoepitopes demonstrates that T cell subsets with T21 show elevated levels of basal IFN signaling and hypersensitivity to IFN-α stimulation. Therefore, these results point to T cell dysregulation associated with IFN hyperactivity as a contributor to autoimmunity in DS.
Keywords: T cells; autoimmunity; inflammation; trisomy 21; type I interferon.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- de Graaf G., Buckley F., Skotko B. G., Estimation of the number of people with Down syndrome in the United States. Genet. Med. 19, 439–447 (2017). - PubMed
-
- Hasle H., Friedman J. M., Olsen J. H., Rasmussen S.A., Low risk of solid tumors in persons with Down syndrome. Genet. Med. 18, 1151–1157 (2016). - PubMed
-
- Madan V., Williams J., Lear J. T., Dermatological manifestations of Down’s syndrome. Clin. Exp. Dermatol. 31, 623–629 (2006). - PubMed
-
- Mårild K., et al. , Down syndrome is associated with elevated risk of celiac disease: A nationwide case-control study. J. Pediatr. 163, 237–242 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

