ILC2s are the predominant source of intestinal ILC-derived IL-10
- PMID: 31699824
- PMCID: PMC7041711
- DOI: 10.1084/jem.20191520
ILC2s are the predominant source of intestinal ILC-derived IL-10
Abstract
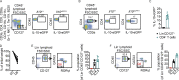
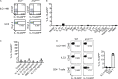
Although innate lymphoid cells (ILCs) functionally analogous to T helper type 1 (Th1), Th2, and Th17 cells are well characterized, an ILC subset strictly equivalent to IL-10-secreting regulatory T cells has only recently been proposed. Here, we report the absence of an intestinal regulatory ILC population distinct from group 1 ILCs (ILC1s), ILC2s, and ILC3s in (1) mice bred in our animal facility; (2) mice from The Jackson Laboratory, Taconic Biosciences, and Charles River Laboratories; and (3) mice subjected to intestinal inflammation. Instead, a low percentage of intestinal ILC2s produced IL-10 at steady state. A screen for putative IL-10 elicitors revealed that IL-2, IL-4, IL-27, IL-10, and neuromedin U (NMU) increased IL-10 production in activated intestinal ILC2s, while TL1A suppressed IL-10 production. Secreted IL-10 further induced IL-10 production in ILC2s through a positive feedback loop. In summary, ILC2s provide an inducible source of IL-10 in the gastrointestinal tract, whereas ILCregs are not a generalizable immune cell population in mice.
© 2019 Bando et al.
Figures



References
-
- Bando J.K., Gilfillan S., Song C., McDonald K.G., Huang S.C., Newberry R.D., Kobayashi Y., Allan D.S.J., Carlyle J.R., Cella M., and Colonna M.. 2018. The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity. 48:1208–1219.e4. 10.1016/j.immuni.2018.04.012 - DOI - PMC - PubMed
-
- Berg D.J., Davidson N., Kühn R., Müller W., Menon S., Holland G., Thompson-Snipes L., Leach M.W., and Rennick D.. 1996. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J. Clin. Invest. 98:1010–1020. 10.1172/JCI118861 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

