Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair
- PMID: 31700156
- PMCID: PMC6838189
- DOI: 10.1038/s41598-019-52637-0
Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair
Abstract
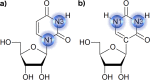
Pseudouridine (Ψ) is the most common chemical modification present in RNA. In general, Ψ increases the thermodynamic stability of RNA. However, the degree of stabilization depends on the sequence and structural context. To explain experimentally observed sequence dependence of the effect of Ψ on the thermodynamic stability of RNA duplexes, we investigated the structure, dynamics and hydration of RNA duplexes with an internal Ψ-A base pair in different nearest-neighbor sequence contexts. The structures of two RNA duplexes containing 5'-GΨC/3'-CAG and 5'-CΨG/3'-GAC motifs were determined using NMR spectroscopy. To gain insight into the effect of Ψ on duplex dynamics and hydration, we performed molecular dynamics (MD) simulations of RNA duplexes with 5'-GΨC/3'-CAG, 5'-CΨG/3'-GAC, 5'-AΨU/3'-UAA and 5'-UΨA/3'-AAU motifs and their unmodified counterparts. Our results showed a subtle impact from Ψ modification on the structure and dynamics of the RNA duplexes studied. The MD simulations confirmed the change in hydration pattern when U is replaced with Ψ. Quantum chemical calculations showed that the replacement of U with Ψ affected the intrinsic stacking energies at the base pair steps depending on the sequence context. The calculated intrinsic stacking energies help to explain the experimentally observed sequence dependent changes in the duplex stability from Ψ modification.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957;227:907–915. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

