Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy
- PMID: 31700181
- PMCID: PMC7938381
- DOI: 10.1038/s41591-019-0639-4
Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy
Abstract
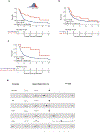
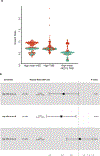
Functional diversity of the highly polymorphic human leukocyte antigen class I (HLA-I) genes underlies successful immunologic control of both infectious disease and cancer. The divergent allele advantage hypothesis dictates that an HLA-I genotype with two alleles with sequences that are more divergent enables presentation of more diverse immunopeptidomes1-3. However, the effect of sequence divergence between HLA-I alleles-a quantifiable measure of HLA-I evolution-on the efficacy of immune checkpoint inhibitor (ICI) treatment for cancer remains unknown. In the present study the germline HLA-I evolutionary divergence (HED) of patients with cancer treated with ICIs was determined by quantifying the physiochemical sequence divergence between HLA-I alleles of each patient's genotype. HED was a strong determinant of survival after treatment with ICIs. Even among patients fully heterozygous at HLA-I, patients with an HED in the upper quartile respond better to ICIs than patients with a low HED. Furthermore, HED strongly impacts the diversity of tumor, viral and self-immunopeptidomes and intratumoral T cell receptor clonality. Similar to tumor mutation burden, HED is a fundamental metric of diversity at the major histocompatibility complex-peptide complex, which dictates ICI efficacy. The data link divergent HLA allele advantage to immunotherapy efficacy and unveil how ICI response relies on the evolved efficiency of HLA-mediated immunity.
Figures



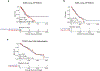





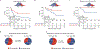

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

